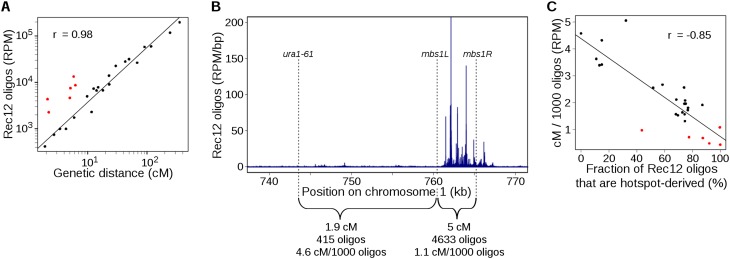
Figure 3.
DSBs in hotspots are much less likely to give rise to crossovers than are DSBs in cold regions. (A) Genetic map length is proportional to Rec12-oligo count, but regions containing strong hotspots behave differently from regions lacking hotspots. Rec12 oligos were summed across intervals with known genetic distances; replicate map lengths were averaged. Sources of map length data are in Supplemental Table S4. The Pearson correlation coefficient r is based on black points; red points are short intervals containing strong DSB hotspots. (B) Different ratios of crossing over to DSB frequency (cM/1000 oligos) at the mbs1 hotspot and an adjacent DSB “cold region.” Genetic distances are from Cromie et al. (2005). (C) DSBs from “cold regions” are more likely to yield a crossover than DSBs in “hot” regions, and the degree of bias in crossover fate correlates with the region’s heat. For each genetic interval in A, the fraction of Rec12 oligos that originated from hotspots was determined (see B for an example). Red points are the outliers in A. See Supplemental Figure S3 for other correlations and estimates of the contribution of different types of intervals to crossover numbers.