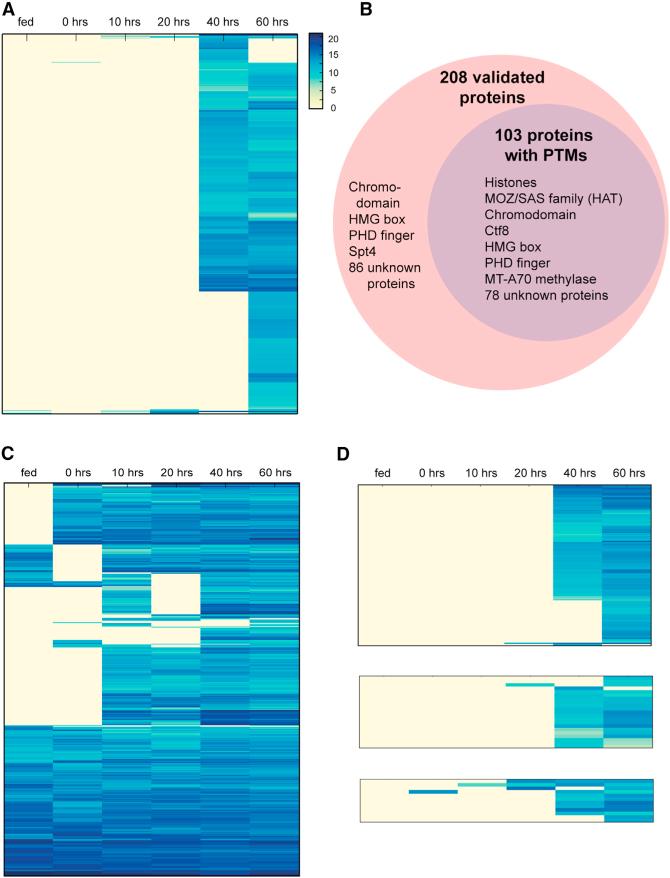
Figure 5. MIC-Limited Genes and Transpo-sons Are Preferentially Expressed during Conjugation, while IES-less Genes Are More Constitutive but Show Universally High Expression during Conjugation.
(A) Clustered expression profile of 810 germline-limited nonrepetitive genes across different time points (vegetative stage (fed); 0, 10, 20, 40, and 60 hr during the conjugating time course). Gene expression levels are represented by log2 (100,000 × normalized RNA-seq counts/coding sequence length).
(B) Mass spectrometry validated 208 MIC-limited genes (outer, pink circle) and 103 were found to contain posttranslational modifications (PTMs) (inner, purple circle). Representative members of each group are shown within the circles.
(C) Clustered expression profiles of 530 IES-less genes.
(D) Clustered expression profiles of MIC-limited transposon-associated genes. Upper: 275 reverse transcriptase and endonuclease domain proteins encoded by LINEs; Middle: 21 Helitron-associated helicases; Lower: 12 DDE_Tnp_IS1595 (ISXO2-like transposase) domain proteins from insertion sequences (ISs).
See also Tables S2, S3, S4, S5, S6, S7, and S8 and Figure S4.