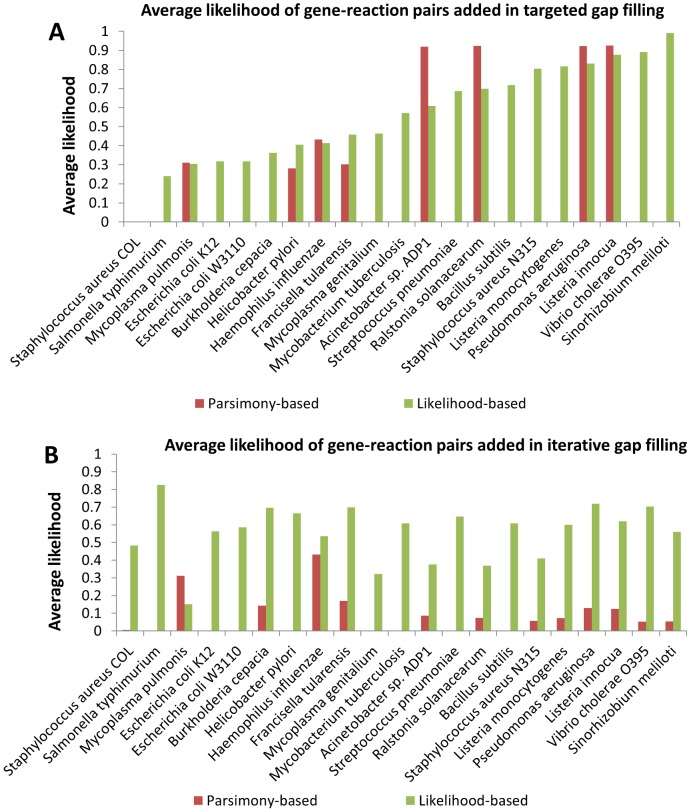
Figure 5. Likelihoods of gene-reaction associations added using likelihood-based and parsimony-based gap filling.
The average likelihood of links between genes and reactions that were added using likelihood-based gap filling tended to be greater than the average likelihood of links resulting from post-processing the parsimony-based gap filling result. Note that it was not greater for all models (e.g., Pseudomonas aeruginosa) because the likelihood-based gap filling approach maximizes likelihood of reactions, not annotations, and as a result picks fewer reactions with 0 likelihood (no predicted gene associations). A) Targeted gap filling result. B) Iterative gap filling result.