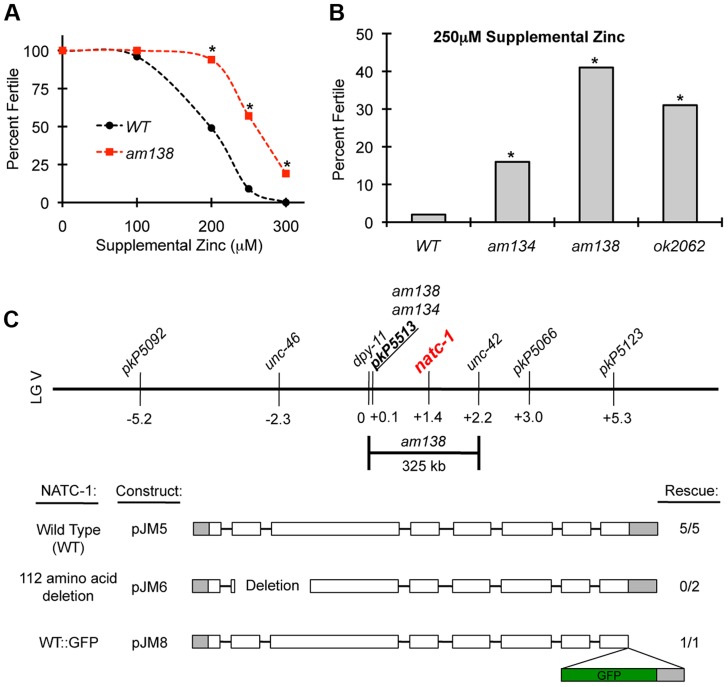
Figure 1. natc-1 mutations cause resistance to high zinc toxicity.
(A) Wild-type (WT) and natc-1(am138) embryos were cultured on NAMM medium with E. coli as a food source and supplemental zinc (µM). Data points indicate the percent of embryos that developed to adulthood and were fertile based on the generation of at least one progeny (N = 25–226 animals). natc-1(am138) animals displayed significant resistance compared to wild-type animals at 200 µM and higher concentrations of supplemental zinc (*, p<0.05). (B) Wild-type, natc-1(am134), natc-1(am138), and natc-1(ok2062) embryos were cultured on NAMM supplemented with 250 µM zinc (N = 45–55). All three natc-1 mutant strains displayed significant resistance to zinc toxicity compared to wild type (*, p<0.05). (C) A genetic map of the center of C. elegans linkage group (LG) V. Loci defined by SNPs and mutations that cause visible phenotypes are named above, and positions in map units are shown below. am134 and am138 displayed tightest linkage to the SNP pkP5513 compared to the other SNP markers [36]. Three-factor mapping experiments positioned am138 between dpy-11 and unc-42, an interval of 325 kb [36] that includes natc-1 (T23B12.4). Transgenic natc-1(am134) animals containing extrachromosomal arrays with plasmid pJM5, which encodes wild-type NATC-1, or plasmid pJM6, which encodes NATC-1 with a 112 amino acid deletion from codon 33–144 resulting in a predicted frameshift in the mutated open reading frame, were assayed for zinc resistance. Transgenic natc-1(am138) animals containing extrachromosomal arrays with plasmid pJM8, which encodes wild-type NATC-1 fused to green fluorescent protein (GFP), were assayed for zinc resistance. Open boxes indicate exons, and shading indicates untranslated regions. The GFP open reading frame is shaded in green. Rescue indicates the number of independently derived transgenic strains in which transgenic siblings displayed significantly reduced resistance to high zinc compared to nontransgenic siblings and the total number of transgenic strains analyzed (Figure S1).