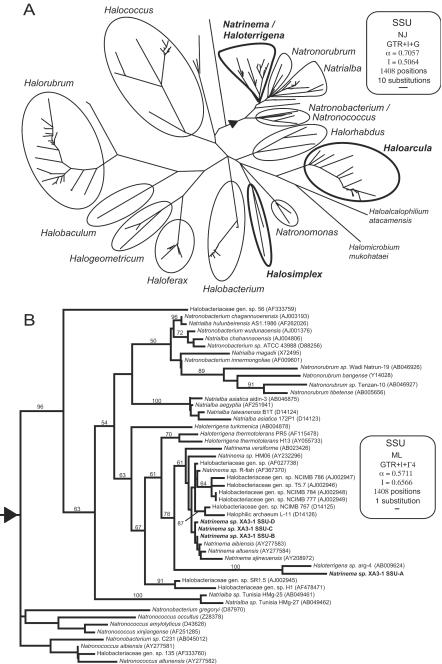
FIG.5.
(A) Best maximum-likelihood distance tree for the SSU gene for the archaeal order Halobacteriales. (B) Best maximum-likelihood tree for the Halobacteriales subgroup in the triangular area in panel A. The evolutionary models and parameters used for the phylogenetic analyses are indicated in the boxes (NJ, neighbor joining; ML, maximum likelihood; GTR, general time reversible; I, proportion of invariable sites; G, gamma-distributed among-site rate variation; α, gamma rate shape parameter alpha; Γ, number of gamma distribution rate categories). The genera in which single organisms contain highly heterogeneous rRNA operons are indicated by boldface type. Bootstrap values were obtained by using a distance maximum-likelihood tree reconstruction method (only values greater than 50% are indicated at the nodes).