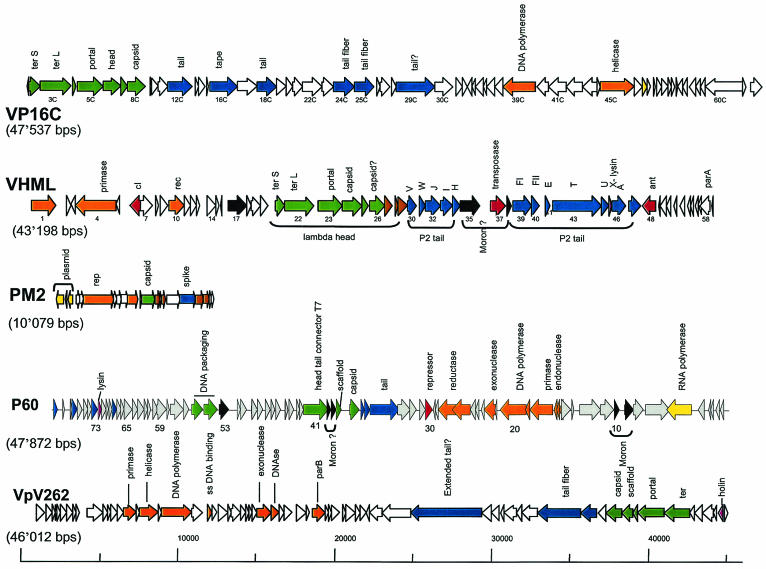
FIG. 3.
Genomic diversity of phages isolated from the ocean. The genome maps of the currently sequenced marine phages are shown. From top to bottom: Vibrio parahaemolyticus phage VP16C resembling dairy Siphoviridae over the structural genes (GenBank accession no. AY328853; AY328852 for the related phage VP16T); Vibrio harveyi phage VHML resembling E. coli siphophage lambda over the head and myophage P2 over the tail genes (no. AY133112); Alteromonas phage PM2, the first lipid-containing bacterial virus attributed to the new phage family Corticoviridae (no. NC_000867); cyanophage P60 infecting Synechococcus sp., which shows a distant E. coli phage T7-like genome organization (no. NC_003390); V. parahaemolyticus phage VpV262, another marine phage with a T7-like genome organization (no. NC_003907). Not shown are a Roseobacter phage SIO1 (no. NC_002519), since it only distantly resembled T7 phages and otherwise nothing else in the database. Also not shown is the broad-host-range vibriophage KVP40 (no. NC_005083) because its 244,835-bp genome is larger than all depicted phage genomes together. KVP40 resembles E. coli myovirus T4 closely over the structural and DNA replication modules but differs by large insertions of DNA lacking database matches. The likely functions of the genes are color coded: green, head; blue, tail; brown, head-to-tail genes (in VP16C) and transmembrane structural genes (in PM2); orange, DNA replication; yellow, transcription; red, lysogeny and recombination; mauve, lysis; black, lysogenic conversion genes.