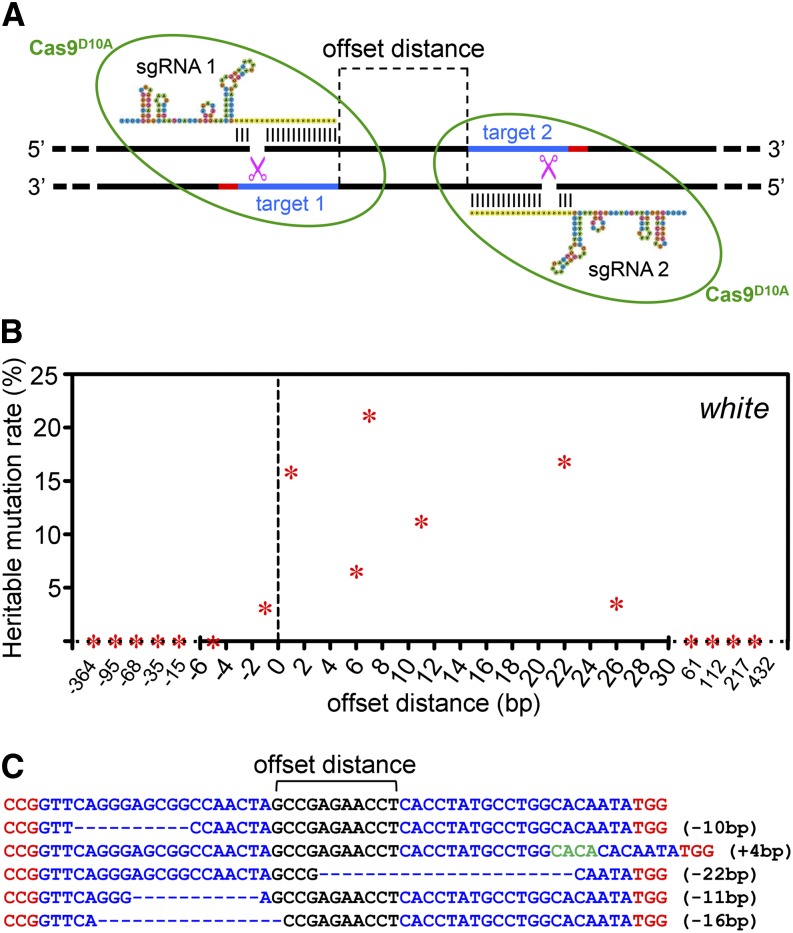
Figure 1.
A pair of sgRNAs targeting close regions in the genome can introduce indel mutations in transgenic Cas9D10A nickase flies. (A) Diagram showing two neighboring Cas9D10A/sgRNA ribonucleoproteins targeting the fly genome. The D10A mutation converts Cas9 into a nickase that targets only the strand complementary to the sgRNA. Each Cas9D10A nickase is shown as a green circle. The sgRNA targets are shown in blue and the PAMs are shown in red. The single-strand cutting sites are shown by the magenta scissors. The offset distance is measured from the PAM-distal end of an sgRNA target to that of the other. If the PAMs are facing outward away from each other as shown in this diagram, then the distance is a positive number. (B) Scatter plot showing the relationship between heritable white mutation rate and sgRNA distance when using offset pairs in the Cas9D10A transgenic flies. Heritable mutation rate is calculated as number of the white-eyed mutant F1 flies divided by total F1 flies screened. (C) Representative sequencing results showing mutations generated with Cas9D10A transgenic flies and a pair of sgRNAs with offset distance of 11 bp. The sgRNA targets are in blue, and the PAMs are shown in red. Mutations with deleted nucleotides are shown with dashed lines, and inserted nucleotides are shown in green.