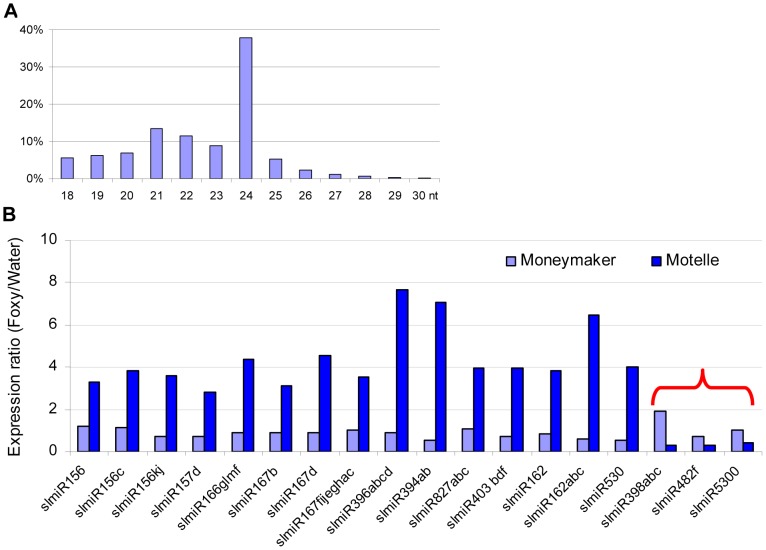
Figure 1. Properties of miRNAs expressed in resistant and susceptible tomato cultivars treated with water or the fungal pathogen Fusarium oxysporum.
In total, four small RNA libraries were subjected to deep sequencing: susceptible tomato cultivar Moneymaker treated with water; Moneymaker treated with Fusarium oxysporum f. sp. lycopersici (F. oxysporum); resistant tomato cultivar Motelle treated with water and Motelle treated with F. oxysporum. The sequence length of small RNAs (A) for the combined data from the four libraries was determined as described in the Materials and Methods. Relative expression levels of known miRNAs (B) were determined by dividing normalized reads for F. oxysporum treatment by those for water treatment for each cultivar. The red bracket indicates the miRNAs that were negatively regulated in Motelle, but not Moneymaker, after F. oxysporum infection. Putative roles/targets of miRNAs in various plant species (information from miRBase.org): miR156, miR156c, miR156kj and miR157d: Squamosa-promoter Binding Protein (SBP)-like transcription factors; miR166glmf: HD-Zip transcription factors, including Phabulosa (PHB) and Phavoluta (PHV) that regulate axillary meristem initiation and leaf development; miR167b, miR167d and miR167fijeghac: Auxin Response Factors (ARF transcription factors); miR396abcd: Growth Regulating Factor (GRF) transcription factors, rhodenase-like proteins, and kinesin-like protein B; miR394ab, F-box proteins; miR827abc: Unknown; miR403bdf: Virus defense; miR162 and miR162abc: Unknown; miR530: Unknown; miR398abc: copper superoxide dismutases and cytochrome C oxidase subunit V; miR482f: NB domain proteins; miR5300, Unknown.