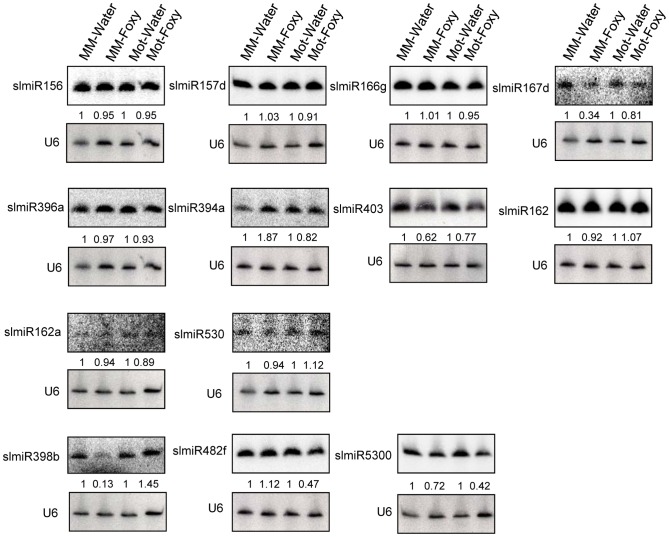
Figure 2. Northern blot analysis of miRNAs.
Oligonucleotide probes were used to quantitate levels of several miRNAs identified during deep sequencing using northern blot analysis. Root total RNA samples (40 µg each) were from Moneymaker treated with water (MM-H2O), Moneymaker infected with F. oxysporum (MM-Foxy), Motelle treated with water (Mot-H2O) and Motelle infected with F. oxysporum (Mot-Foxy). U6 RNA served as a loading control for each blot. Blots were imaged using a Phosphorimager and miRNA species quantitated using Imagequant software, with normalization to the amount of U6 RNA. The numbers below each blot indicate the amount of miRNA in each sample relative to the corresponding water-treated control. Note that due to significant sequence homology, slmiR482 subfamily members cannot be quantitatively distinguished from one another using northern analysis.