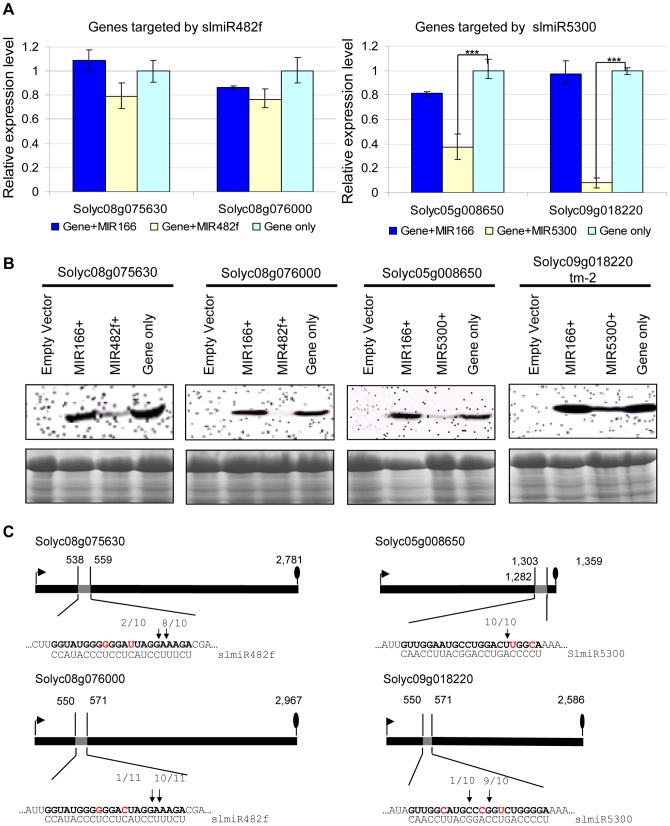
Figure 5. Co-expression of slmiRNAs and predicted targets in Nicotiana benthamiana leaves.
A. Levels of target mRNAs. qRT-PCR was used to determine relative levels of predicted tomato mRNAs in N. benthamiana leaves expressing only empty vector; target mRNA and a control miRNA (slmiR166); target mRNA and the appropriate miRNA (slmiR482f or slmiR5300); or target mRNA and empty vector. Values were normalized to N. benthamiana actin. Errors are expressed as the standard error. *** indicates significant differences when compared to the corresponding control plants in the same treatments at p<0.001. B. Target protein levels. Total protein isolated from the samples in (A) was electrophoresed on SDS-PAGE gels and blotted onto nitrocellulose membranes. A FLAG antiserum was used to detect the tagged target proteins during western analysis as described in the Materials and Methods (top panels). A duplicate gel was Coomassie-stained and used as a loading control (bottom panels). Similar results were obtained for two biological replicates. C. Diagrammatic representation of target mRNA cleavage sites determined using 5′RACE. Thick black lines represent open reading frames (nucleotide numbering begins at the start codon indicated by the black arrow), and the putative miRNA interaction site is shown as a gray box, with the nucleotide position within the ORF indicated. The nucleotides shown in red in each mRNA target are mismatches with the corresponding miRNA. The number of sequenced 5′RACE clones corresponding to each site is indicated by vertical arrowheads.