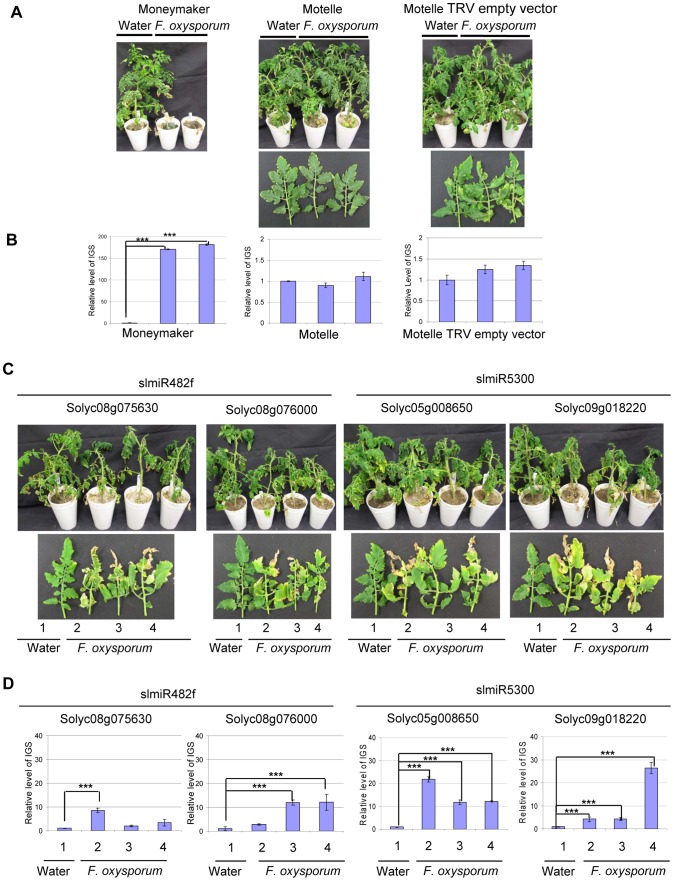
Figure 7. Targets of slmiR482f and slmiR5300 are required for F. oxysporum defense in tomato.
All control and VIGS plants were cultivated together in the same growth chamber. Similar results were obtained in three biological replicates. Errors are expressed as standard error. *** indicate significant differences when compared to the corresponding control plants in the same treatments at p<0.001. A. Phenotype of control plants infected by Fusarium oxysporum . Two-week-old seedlings of the indicated cultivars were treated with water or F. oxysporum and photographed four weeks later. Cultivars were Moneymaker (left panel), Motelle (center panel) and Motelle infected with TRV empty vector (right panel). B. The relative level of F. oxysporum in control plants. Leaves were collected from water or F. oxysporum-treated plants and genomic DNA isolated. qPCR of the Intergenic Spacer region (IGS) of F. oxysporum was used to measure relative fungal cell loads in planta. Note the different y-axis scale for Moneymaker (left graph) vs. Motelle (center and right graphs). C. Phenotypes of VIGS plants after infection with F. oxysporum . Motelle tomato plants were infiltrated with constructs carrying vectors that would lead to VIGS of a predicted target gene. Results for the two targets of slmiR482f are shown to the left, while the two for slmiR5300 are shown to the right. Four weeks after initiation of VIGS, roots of tomato plants were infected with F. oxysporum or with water (controls). Phenotypes were scored four weeks later. A water-treated control VIGS plant or a leaf is shown on the left of each panel, with three plants or leaves from F. oxysporum-infected VIGS plants to the right. D. qPCR to determine relative levels of F. oxysporum cells in leaves of VIGS plants. qPCR of the F. oxysporum rRNA intergenic spacer region was performed on genomic DNA isolated from leaves of water or F. oxysporum- infected plants as described in (B).