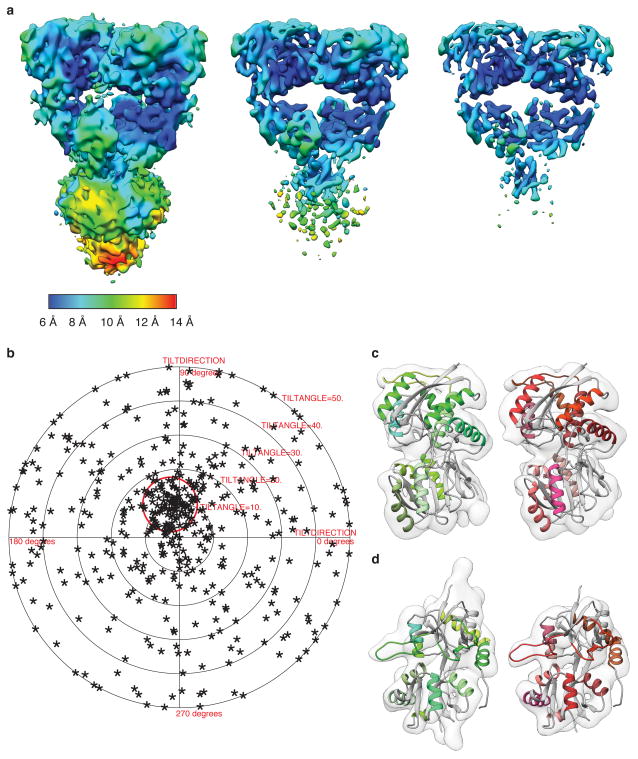
Extended Data Figure 8. Resolution of the desensitized GluK2 density map.
a, GluK2 desensitized state map shown at increasing contour levels from left to right, to better highlight selected secondary structural features. b, Validation of density map using tilt-pair parameter plot. The spread in orientational assignments around the known goniometer settings is within ~ 25° for > 60 % of the selected particle pairs, with clear clustering observed at the expected location, centered at a distance of 10° from the origin. c, Distal (left) and proximal (right) ATD subunits fit with the corresponding X-ray coordinates (PDB ID: 3H6G). d, Proximal (left) and distal (right) LBD subunits fit with the corresponding X-ray coordinates for glutamate-bound GluK2 LBD monomers (PDB ID: 3G3F). The close similarity in density maps for the individual ATD and LBD monomers of distal and proximal domains that are unrelated by computationally imposed C2 symmetry shows that the LBD monomers move largely as rigid bodies and that the structural changes that occur with desensitization can be described adequately by rigid body movements of the ATD and LBD monomers.