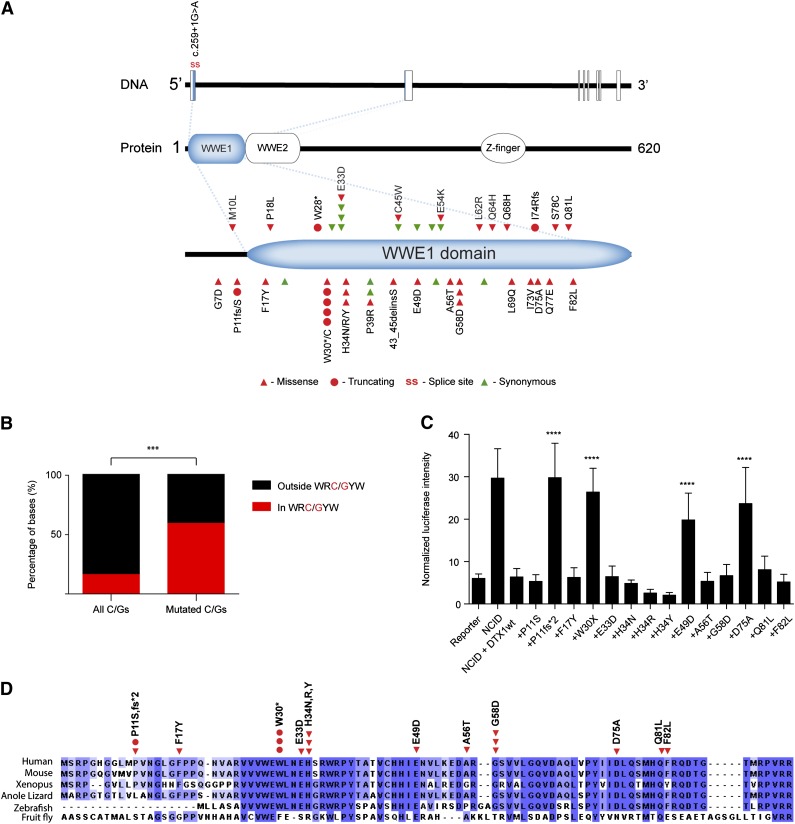
Figure 6.
Analysis of DTX1 mutations. (A) Diagram displaying mutations identified in DTX1. Mutations were restricted to the first exon of the gene that encodes for most of the WWE1 domain of DTX1. (B) Distribution of DTX1 mutations by AID targeting hotspot. Distribution of all and mutated cytosines and guanines by WRC/GYW domains in the first exon of DTX1. In spite of the majority of cytosines and guanines being located outside WRC/GYW domains, most mutations were encountered at this AID target motif. (C) Functional characterization of DTX1 mutants. Functional characterization of mutations identified in DTX1. Cotransfection of constructs containing the NCID, DTX1 wild-type, or mutant constructs, and a CSL-responsive luciferase reporter, was performed in U2OS cells. A control pRL-TK plasmid was also cotransfected to allow internal normalization of luciferase activity. The capacity to inhibit an NCID-triggered response by DTX1 was diminished in 4 mutants (p.P11fs*2, p.W30*, p.E49D, and p.D75A). Mann-Whitney U tests were performed to compare the firefly luciferase intensity in wild-type and mutant DTX1 transfected cells (***P < .001). The means and standard deviations of 3 independent experiments are displayed, each containing 3 replicates. (D) Multispecies alignment of the N-terminal portion of DTX1, encoded by the first exon of the gene. The mutations tested in the Notch reporter assay are displayed. Multiple sequence alignment was performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and visualized with Jalview.37