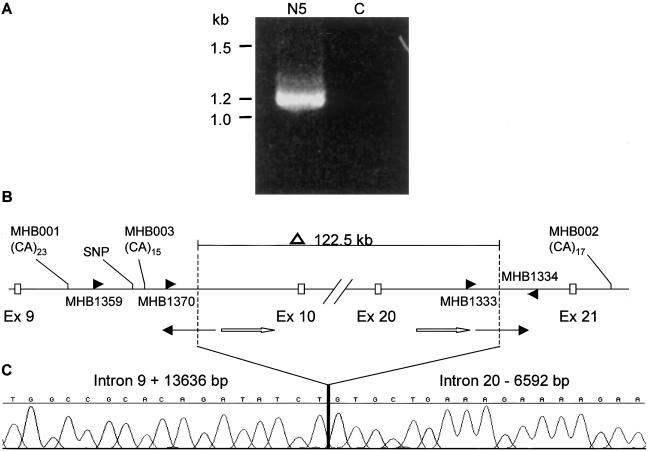
Figure 2.
A, Breakpoint-spanning fragment amplified by primers MHB1359 and MHB1334. In a Navajo individual with OCA2 (N5), a 1.2-kb fragment amplified, whereas in the control subject (C), no fragment was detected. B, Sequences flanking the breakpoints. The breakpoints are represented as vertical dashed lines, which are located at positions intron 9 + 13636 bp and intron 20 −6592 bp. As shown above, 122.5 kb of the P gene is deleted, including exons 10–20. The primers, MHB1359/1334, were used to amplify across the breakpoints (panel A) and the three primers, MHB1370/1333/1334, were used for detecting carriers. Microsatellite markers MHB001, MHB002, MHB003, and the SNP were used in haplotype analysis. MHB001, located 8.6 kb upstream of the left-hand breakpoint, consists of a perfect (CA)23 repeat (corresponding to the 466-bp allele). The SNP (T/C) is 703 bp upstream of the left-hand breakpoint. MHB003 is 118 bp upstream of the left-hand breakpoint and consists of compound repeat (CA)10(TA)1(CA)4. MHB002 is 20 kb downstream of the deletion and consists of a perfect (CA)17 repeat (corresponding to the 370-bp allele). At the breakpoint juncture, two LINE sequences (thin arrows) are at each end and are oriented in opposite directions. The two homologous LINE sequences (white arrows) are 3,032 bp downstream of the left-hand breakpoint and 1,481 bp upstream of right-hand breakpoint. The homologous LINE sequences share 85% identity in a 745-bp stretch and are oriented in the same direction. C, Chromatograph of the sequence flanking the breakpoints.