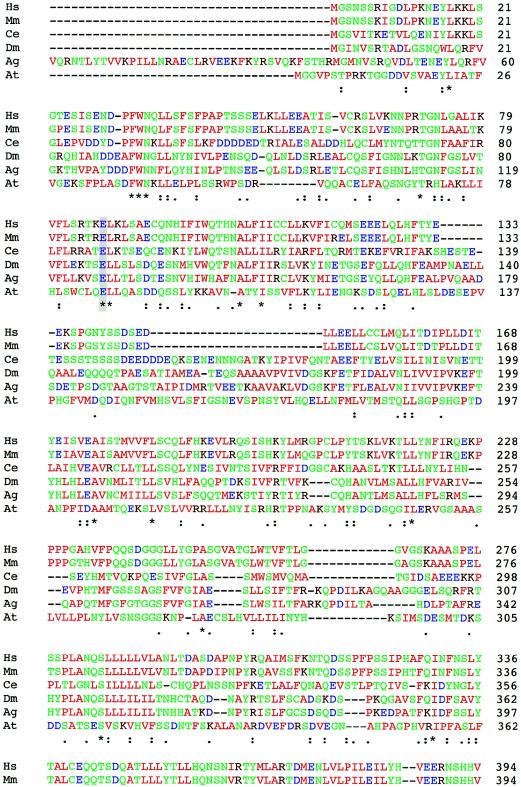
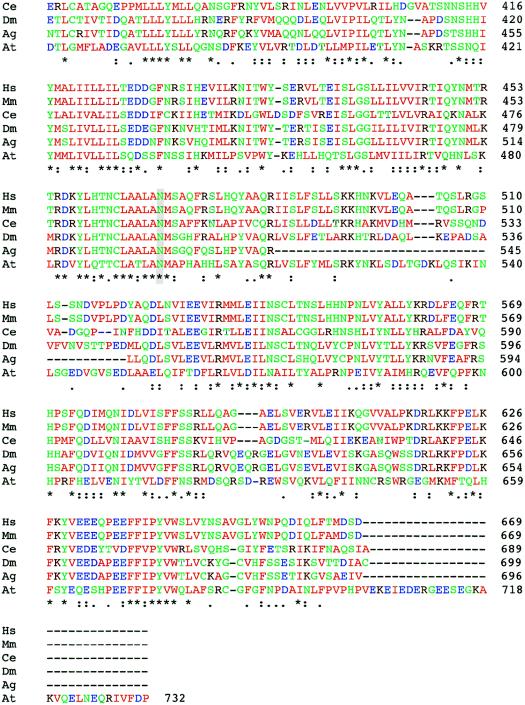
Figure 3.
Comparison of the protein sequences of FLJ90130 and homologous proteins in other species. ClustalW was used for the alignment. The species from which each sequence is derived is shown on the left (M. musculus [Mm], D. melanogaster [Dm], A. gambiae [Ag], C. elegans [Ce], and A. thaliana [At]), and the amino acid residue numbers are shown on the right. Colors are used to indicate classes of amino acid residues, including hydrophobic and aromatic (green), acidic (blue), basic (dark red), and hydrophilic (green) residues. Symbols below the amino acid sequences indicate identity (*), presence of conserved substitutions (:), or presence of semiconserved substitutions (.). A gray background highlights the residues changed by the two identified missense mutations, E87 and N469.