FIGURE 1.
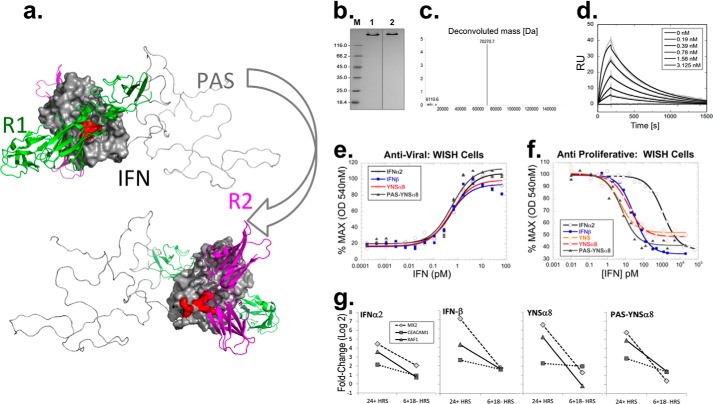
Development and evaluation of PAS-YNSα8. a, PAS-YNSα8 was depicted using PyMOL, also showing IFNAR1, IFNAR2, and the PAS tag (here with 200 residues in one exemplary random coil conformation) as cartoons in green, magenta, and gray, respectively, using two orientations rotated by 180°. Mutations H57Y, E58N, and Q61S (YNS) and E159K, S160R, and R162K (α8-tail) are depicted on the IFN surface facing IFNAR1 and IFNAR2, respectively. b, analysis of PAS-YNSα8, purified from the periplasmic cell fraction of E. coli, by Coomassie-stained 12% SDS-PAGE under reducing (lane 1) and nonreducing (lane 2) conditions. Lane M, protein size marker. c, ESI-MS analysis of PAS-YNSα8, confirming monodisperse composition and the calculated average molecular mass of 70,271.8 Da (ExPASy ProtParam tool). x-axis: mass (Da); y-axis: counts (×10,000). d, representative real time SPR analysis of PAS-YNSα8 binding to the soluble human IFNAR2-Fc chimera immobilized at ΔRU = 200–250 on a Xantec CMDP sensor chip measured on a BIAcore 2000 instrument and fitted to a 1:1 Langmuir model. The resulting kinetic and affinity parameters are listed in Table 1. e and f, anti-viral (e) and antiproliferative (f) dose-response curves in human WISH cells for different IFN subtypes. g, transcript expression levels of representative IFN-I response genes after stimulation with 1 pm of different human IFN-Is. The cells were exposed to the different IFNs for 24 or 6 h (followed by 18 h in IFN-free medium) before harvest and gene transcript analysis.