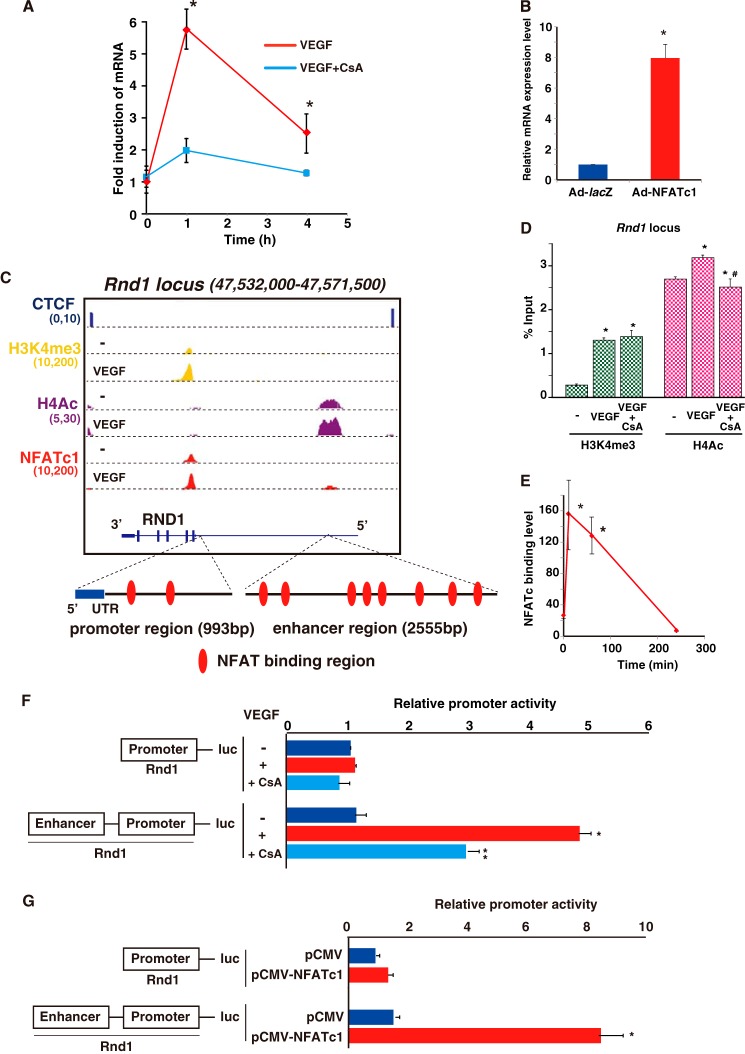
FIGURE 6.
NFATc1 binds and transactivates RND1 in VEGF-treated endothelial cells. A, real-time qPCR analysis of RND1 expression in HUVEC treated with VEGF alone (red) or treated with VEGF plus CsA (blue) at the indicated time points *, p < 0.01 compared with VEGF plus CsA (n = 3). B, RND1 expression in HUVEC infected with Ad-NFATc1 or Ad-lacZ for 24 h (*, p < 0.001 compared with Ad-lacZ) (n = 3). C, genome browser view around the Rnd1 gene. ChIP signals in the presence or absence of VEGF are shown with NFATc1 (red), H3K4me3 (yellow), and H4Ac (purple). Insulator CTCF bindings are shown in blue. The number below each ChIP-seq indicates the threshold window on the IGB software. D, HUVEC were treated with VEGF for 1 h. ChIP was performed with antibodies to H3K4me3 (green) and H4Ac (pink). Subsequently, qPCR was performed using primers specific to the Rnd1 target locus. The fold enrichment was compared with non-immunoprecipitated chromatin (percent input). Data are mean ± S.E. from three independent experiments. *, p < 0.05 compared with to without VEGF; #, p < 0.05 compared with to without CsA (n = 3). E, ChIP-qPCR with NFATc1 antibody on the Rnd1 locus in HUVEC treated with VEGF at the indicated time points. *, p < 0.001 compared with 0 min (n = 3). F, HUVEC were transfected with Rnd1-luc or Rnd1 (NFAT mut)-luc, serum-starved, preincubated with 1 μm CsA or mock control, and treated with VEGF for 4 h. Data are mean ± S.D. of luciferase light units obtained from six independent experiments. *, p < 0.001 compared with to without VEGF; **, p < 0.01 compared with to without CsA. G, RND1-luc and the NFATc1 expression plasmid (pCMV-NFATc1) were transiently cotransfected into COS7 cells. Two days later, cell lysates were assayed for reporter activity. Data are mean ± S.D. of relative expression compared with the mock control (pCMV). *, p < 0.001 compared with the control (n = 6).