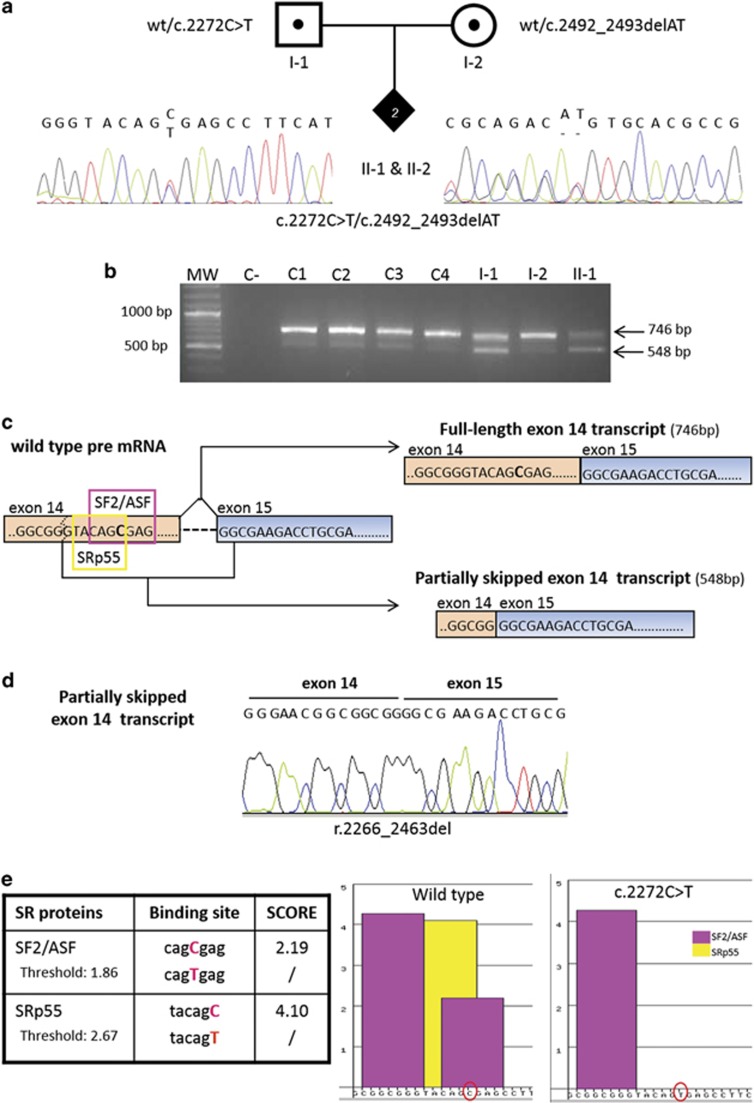
Figure 2.
DNA and RNA characterisation of RECQL4 mutations in the index family. (a) Pedigree of the family with two siblings with Rothmund–Thomson syndrome (filled diamond) found to be compound heterozygotes for the paternal c.2272C>T and the maternal c.2492_2493delAT mutations shown by the electropherograms. (b) cDNA analysis of the RECQL4 region spanning exons 11–15 showing the expected 746 bp amplicon as well as a shorter, 548 bp, band observed in all samples but with a higher intensity in the affected daughter II-1 and carrier father I-1. MW, molecular weight; C−, negative control; C1, C2, C3, C4, control samples; I-1, father; I-2, mother; II-1, index case. (c) Diagram based on sequence analysis of the two gel bands in (b) showing the canonical and alternative splicing of IVS14. Two different transcripts originate from the RECQL4 pre-mRNA (right): the full-length exon 14 transcript of 746 bp (upper right) and the partially skipped exon 14 transcript of 548 bp (lower right). The two different SR proteins recognising the ESE motifs in the region of exon 14 including the mutation site (C>T in bold) are framed in purple (SF2/ASF) and yellow (SRp55) on RECQL4 pre-mRNA. The dashed line indicates the exonic cryptic splice site leading to the partially skipped exon 14 transcript. (d) Sequence electropherograms showing the physiological partially skipped exon 14 transcript (r.2266_2463del). (e) The table on the left shows the two SR proteins recognising the ESE motifs in RECQL4 exon 14 close to the c.2272 mutation site (in red) and their scores in the presence of the wild-type and c.2272C>T sequence. The diagram on the right shows ESE binding site(s) of SF2/ASF (purple bars) and SRp55 (yellow bar) proteins on the RECQL4 wild-type and mutant sequences reported on the x axis (C and T red encircled). The C>T mutation abrogates one of the binding sites for SF2/ASF and the binding site for SRp55.