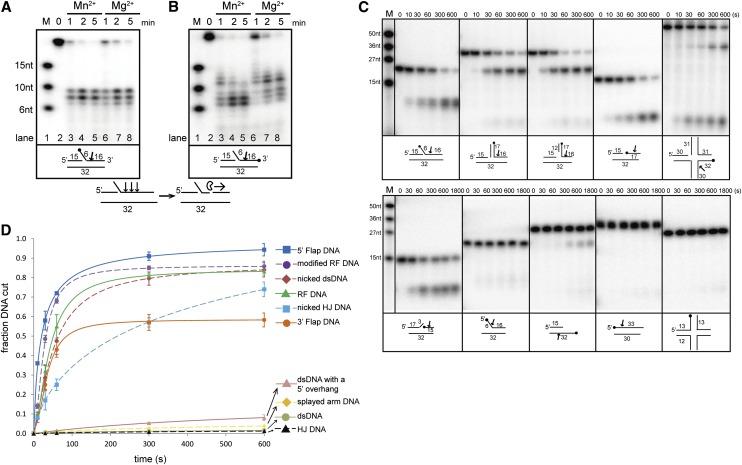
Figure 1.
Biochemical characterization and overall structure of PaFAN1 bound to DNA. Endonuclease (A) and exonuclease (B) activities of PaFAN1 in the presence of Mn2+ and Mg2+ ions. The labeled site is marked with a filled circle. Although PaFAN1 exhibited nuclease activity in the presence of Mg2+ or Mn2+, nucleolytic activity was more efficient in the presence of Mn2+, especially when FAN1 mutant proteins were used (see Fig. 4A; Supplemental Fig. 6). PaFAN1 incises the third, fourth, or fifth phosphate group (arrow) downstream from the junction between the ssDNA and dsDNA and cleaves further in the 3′ direction. (C) Nuclease activities of PaFAN1 toward various substrates in the presence of Mn2+ ions. (Top) 5′ flap (lanes 2–7), RF (lanes 8–13), modified RF (lanes 14–19), nicked dsDNA (lanes 20–25), and nicked HJ (lanes 26–31). (Bottom) 3′ flap (lanes 2–7), splayed arm (lanes 8–13), dsDNA with a 5′ overhang (lanes 14–19), dsDNA (lanes 20–25), and intact HJ (lanes 26–31). See Supplemental Figure 8 for detailed substrate structures. (D) The cleavage products shown in C have been quantified. The fraction of DNA cut is the ratio of the relevant cleavage product to total DNA (cleaved plus uncleaved DNA). Data were plotted as a function of time using Origin software. From these data, we calculated observed rates of cleavage of 5′ flap DNA (>0.046 sec−1; blue square and line), modified RF (>0.043 sec−1; purple circle and dash), nicked dsDNA (>0.042 sec−1; brown diamond and dash), RF (>0.040 sec−1; green triangle and line), 3′ flap DNA (>0.016 sec−1; orange circle and line), nicked HJ (>0.014 sec−1; cyan square and dash), dsDNA with a 5′ overhang (>0.0015 sec−1; pink triangle and line), splayed-arm DNA (>0.0014 sec−1; yellow diamond and dash), dsDNA (>0.0002 sec−1; gray circle and line), and intact HJ (>0.0002 sec−1; black triangle and dash).