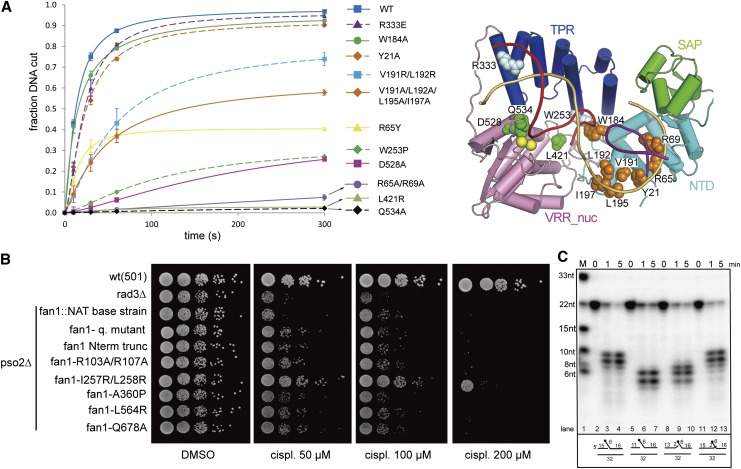
Figure 4.
Endonuclease and exonuclease activities of wild-type and mutant PaFAN1. (A) The 5′ flap DNA nuclease activities of various PaFAN1 mutants were examined at different times (10–300 sec) and quantified. (Blue square and line) Wild type (WT); (purple triangle and dash) R333E; (green circle and line) W184A; (orange diamond and dash) Y21A; (cyan square and dash) V191R/L192R; (orange diamond and line) V191R/L192R/L195A/I197A; (yellow triangle and line) R65Y; (green diamond and dash) W253P; (magenta square and line) D528A; (purple circle and line) R65A/R69A; (gray triangle and line) L421R; (black diamond and dash) Q534A. Assay conditions were the same as those in Figure 1C. The 5′ flap DNA was incubated with the indicated FAN1 protein in the presence of Mn2+ ions. Cleavage products were resolved by denaturing polyacrylamide gel electrophoresis and visualized by phosphoimaging. A schematic diagram of the positions of the mutated residues is shown in the right panel. NTD residues are shown as orange spheres, TPR residues are pale cyan spheres, and VRR nuc residues are green spheres. (B) Sensitivity of point mutations in the conserved residues of S. pombe Fan1 to cisplatin (cispl). A pso2-deleted background was used to compare the effect of the mutations on the hypersensitive double-mutant fan1-deleted pso2-deleted strain. Logarithmically growing cultures were serially diluted and spotted onto a YEA-DMSO control plate and YEA plates containing the cross-linking agent cisplatin at the concentrations indicated. A checkpoint-deficient rad3-deleted strain was used as a hypersensitive control to test the efficacy of the cisplatin used. (C) To identify the reference point of the incision, PaFAN1 was incubated with 5′ flap substrates with various gap sizes. Detailed substrate structures are shown in Supplemental Figure 8.