Abstract
The publication of the first high-resolution crystal structure of a eukaryotic Cys-loop receptor, GluClα, has provided valuable structural information on this important class of ligand-gated ion channels (LGIC). However, limited functional data exist for the GluCl receptors. Before applying the structural insights from GluCl to mammalian Cys-loop receptors such as nicotinic acetylcholine and GABA receptors, it is important to ensure that established functional features of mammalian Cys-loop receptors are present in the more distantly related GluCl receptors. Here, we seek to identify ligand-binding interactions that are generally associated with Cys-loop receptors, including the frequently observed cation−π interaction. Our studies were performed on the highly homologous GluClβ receptor, because GluClα is not activated by glutamate in Xenopus laevis oocytes. Mutagenesis of the signal peptide and pore lining helix was performed to enhance functional expression and sensitivity to applied ligand, respectively. Conventional and unnatural amino acid mutagenesis indicate a strong cation−π interaction between Y206 and the protonated amine of glutamate, as well as other important ionic and hydrogen bond interactions between the ligand and the binding site, consistent with the crystal structure.
The Cys-loop receptors are a family of pentameric ligand-gated ion channels involved in fast synaptic transmission. They are the target of a variety of native neurotransmitters, along with therapeutics targeting, among others, Parkinson’s disease, Alzheimer’s disease, schizophrenia, pain, and nicotine addiction in humans.1−3 Among vertebrates, members of this family include the excitatory, cation-permeable, nicotinic acetylcholine receptors (nAChRs) and 5-HT3A receptors, and the inhibitory, anion-permeable, GABAA/C, and glycine receptors. These receptors possess conserved structural elements, including an extracellular ligand binding domain, four transmembrane helices of which one, M2, forms the channel pore, and the eponymous disulfide loop in the extracellular domain.
Until recently, structural models of Cys-loop receptors were based on the low resolution cryo-EM structures of the Torpedo marmorata nAChR4,5 or the high resolution X-ray crystal structures of acetylcholine binding proteins,6 soluble proteins homologous to the extracellular, ligand-binding domain of the full length receptor. However, recent developments in membrane protein crystallography have led to a wealth of new high-resolution structures including two homologous prokaryotic ligand-gated ion channels, ELIC and GLIC.7−9 Most recently, a structure has been reported for the first eukaryotic Cys-loop receptor, GluClα, from Caenorhabditis elegans.10
GluCl represents a true, eukaryotic Cys-loop receptor, although no vertebrate GluCl has been identified. It is an anion-permeable channel, and it displays 34% sequence identity to its closest human homologue, the α1 glycine receptor. The GluClα receptor was crystallized in the presence of the native orthosteric ligand, glutamate, as well as the positive allosteric modulator, ivermectin, an antiparasitic used to prevent nematode infections in humans (thus preventing river blindness) and in animals.3,11−13 Both ligands were shown to bind at the interface of two subunits. At the orthosteric glutamate site, the primary (+) face of one subunit contributes loops A–C, and the complementary (−) face of an adjacent subunit contributes loops D–F, as is seen in other Cys-loop receptors.
While the GluClα structure represents a milestone in the study of Cys-loop receptors, relatively few functional studies on this receptor have been reported. In contrast, a large number of wide-ranging functional studies of mammalian Cys-loop receptors have been reported.1,14,15 In our own laboratories, we have developed a series of functional probes based on unnatural amino acid mutagenesis that provide high precision information on drug-receptor interactions and protein structural features. These include a cation−π interaction between the protonated amine of endogenous ligands and one of several conserved aromatic amino acids in the binding site, which has been demonstrated in all Cys-loop receptors studied to date.16 In addition, several hydrogen bonding interactions, including hydrogen bonds to the protein backbone, have been established to be important in several Cys-loop receptors.
In the present work, we set out to determine whether key structural features of the C. elegans GluClα receptor have functional consequences that parallel those seen in mammalian Cys-loop receptors. Mutagenesis was performed on the closely related GluClβ receptor, because the GluClα receptor is not activated by glutamate in the absence of ivermectin in Xenopus laevis oocytes.17 Every ligand-binding residue considered here is conserved in the two forms. To facilitate these studies, the receptor had to be optimized for expression in Xenopus oocytes by altering the signal peptide and by introducing a mutation in the pore-lining region. Our studies find functional significance for many of the key features of the GluCl receptor structure and also highlight the challenges of probing binding interactions of invertebrate receptors.
Results and Discussion
Mutagenesis of Pore Lining Residues to Increase the Sensitivity of GluClβ to Glutamate
Similar to many nAChRs, the GluClβ homomeric receptor gives small current responses to applied glutamate and demonstrates a relatively high wild type EC50 of 0.35 mM. The high EC50 value is problematic for measuring loss-of-function mutations for many reasons. Glutamate is not soluble at concentrations higher than 50 mM in buffered solution, and while a hydrochloride salt increases this solubility, it also dramatically increases the osmolarity of the solution to several times that of frog Ringer solution.21 Also, at high concentrations of glutamate, currents are observed in uninjected oocytes, which may be due to nonspecific activation of endogenous receptors or osmotic response. These currents, while small, complicate measurement of low-expressing mutants. Also, in high expressing mutants, rapid desensitization of GluClβ occurs at concentrations greater than 100 mM, prohibiting further measurements. Therefore, to enable measurement of low expressing and large loss-of-function mutants, it is important to lower the EC50 of the wild type receptor.
Mutation of pore lining residues on the M2 helix of Cys-loop receptors has had dramatic effects on ligand sensitivity,22−24 open channel duration,22,25 and receptor desensitization23,26 in many studies, thereby facilitating measurement of low expressing and large loss-of-function mutations.24,27−29 The GluClα crystal structure (PDB ID: 3RIF) indicates that the pore-lining residues are at the −2′, −1′, 2′, 6′, 9′, 10′, 13′, 17′, and 20′ positions in the M2 helix (Supporting Information (SI) Figure S1). These residues physically shape the channel and influence the conductance and the equilibrium between inactive and active conformations through the presence or absence of steric bulk and polarity of the side chains.30 In this study, to modulate these properties of the receptor, the pore-lining residues were mutated to amino acids of varying size and polarity (Table 1). Constitutive activity, a complication in many studies,30 was only observed in mutations at the 9′ site, whereas mutations to the other residues had varied effects on receptor EC50 and current across the membrane. Of the mutations tested, the T6′S mutation led to the greatest increase in receptor sensitivity (44-fold), followed by mutation of the T10′ residue to either alanine or serine (27 and 32-fold, respectively) and the T13′A mutation (9-fold). The T6′A and T13′S mutants were not functional. These results suggest that smaller side chains near the pore constriction facilitate channel opening, and that while polarity of the 6′ side chain is required, it is not important at the 10′ or 13′ positions. The double mutant T6′S/T10′S gave large constitutive currents and small agonist-induced currents, indicative of constitutive activity, as was observed in the L9′A and L9′F mutants.
Table 1. GluClβ M2 Helix Mutations.
| mutation | EC50 (mM) | Hill | n | Imax (μA) | fold shift |
|---|---|---|---|---|---|
| WT | 0.35 ± 0.01 | 2.5 ± 0.2 | 20 | 0.4–4 | |
| AG-2′,-1′PA | 0.46 ± 0.01 | 1.8 ± 0.1 | 10 | 12–66 | 1.3 |
| A2′S | 0.63 ± 0.01 | 2.0 ± 0.1 | 12 | 0.6–6 | 1.8 |
| A2′T | no response | ||||
| T6′A | very low response | ||||
| T6′S | 0.0080 ± 0.0005 | 2.4 ± 0.3 | 27 | 0.06–8 | 1/44 |
| L9′A | constitutively active | ||||
| L9′F | constitutively active | ||||
| L9′V | no response | ||||
| T10′A | 0.013 ± 0.001 | 2.8 ± 0.1 | 14 | 2–28 | 1/27 |
| T10′S | 0.011 ± 0.001 | 2.5 ± 0.1 | 14 | 0.3–4 | 1/32 |
| T13′A | 0.038 ± 0.001 | 2.8 ± 0.1 | 11 | 0.1–6 | 1/9 |
| T13′S | no response | ||||
| A17′G | >5 | 10 | |||
| A17′S | 0.23 ± 0.01 | 2.5 ± 0.1 | 10 | 0.6–12 | 1/1.5 |
| A17′T | 0.058 ± 0.001 | 2.8 ± 0.1 | 13 | 0.3–13 | 1/6 |
| A20′S | no response | ||||
| A20′T | 0.11 ± 0.02 | 1.7 ± 0.1 | 14 | 0.5–48 | 1/3 |
| T6′S/T10′S | constitutive activity, some glutamate response | ||||
To test whether the T6′S mutation affects evaluation of other side chain substitutions, a distal loss-of-function mutation was incorporated into the wild type and T6′S containing receptors for comparison (SI Table S1). This mutation, Y206F, is located in the extracellular domain of the receptor at the agonist binding site and disrupts an important hydrogen bond between the bound glutamate and a nearby threonine side chain, T203 (see below). The loss-of-function observed in the T6′S/Y206F mutant was 12.5-fold, compared to 12.3-fold in the single Y206F mutant. These similar values indicate that the T6′S mutation provides a constant multiplicative factor on other mutations and should not affect measurement of binding site interactions. No change in maximal agonist-induced currents was observed in T6′S-containing receptors. Because of the large increase in glutamate sensitivity and a demonstrated lack of coupling between the T6′S mutation and the ligand binding site, we concluded that incorporation of T6′S as a background mutation is appropriate for measuring large loss-of-function mutations in the GluClβ receptor.
Of the other mutations tested, only small shifts in EC50 were observed. Mutagenesis of the −2′ and −1′ residues to match the GluClα receptor sequence (AG to PA) resulted in no shift of EC50 but a large increase in agonist-induced current. It is also interesting to note that while the A2′S mutation had minimal effect on receptor function, the A2′T mutation abolished glutamate-induced currents. The converse mutation in GluClα, T2′A, was previously shown to restore glutamate sensitivity to an otherwise glutamate-insensitive channel.17 It is therefore likely that the 2′ residue plays an important role in coupling ligand binding to receptor activation.
Optimization of Receptor Expression
It has been shown previously that surface expression of the GluClβ homomeric receptor in HEK293 cells is low relative to the homomeric GluClα and the heteromeric GluClαβ receptors, because it is retained in the endoplasmic reticulum (ER) more so than other GluCl receptors.24 We observe this functionally in X. laevis oocytes, with approximately 10-fold enhanced currents of the heteromer relative to the GluClβ homomeric receptor (data not shown). Data for the homomeric GluClα receptor are not available, because this receptor is not gated by glutamate in X. laevis oocytes.17 The relatively low receptor expression of GluClβ does not affect studies of wild type GluClβ or conventional mutants. However, expression becomes an issue when attempting to incorporate unnatural amino acids by nonsense suppression, which often leads to lower expression levels. In these cases, it becomes problematic to distinguish between a receptor that does not respond to the applied glutamate concentrations and a receptor that is expressed too poorly to detect. To avoid this issue, we sought to increase receptor expression without affecting the receptor’s function.
To enhance functional surface expression of GluClβ receptors, point mutants or chimeras with GluClα segments were made in regions of the receptor previously shown to affect Cys-loop receptor trafficking and expression levels (SI Figure S2).31−37 These regions include the signal peptide, the C-terminus, and the intracellular M3-M4 loop, which contains two RxR putative ER retention motifs in the GluClβ receptor. Chimeras were made with GluClα because it does not substantially accumulate in the ER and because of the high similarity between the receptors, which minimizes the chance of affecting receptor function. Of the mutations and chimeras examined, only GluClβ with the α signal peptide and the RSR/RRR_AAA mutant (replacement of all six residues in the two ER retention motifs with alanine) gave enhanced expression relative to the wild type receptor (SI Figure S3 and Table S2). Conversely, introduction of a putative ER export motif (LxxLE) in the M3-M4 loop decreased expression, and the M3-M4 loop and α C-terminus chimeras resulted in nonfunctional receptors. Mutation of either RxR motif singly to AAA had no effect on receptor expression and replacement of the GluClβ α1 helix with the corresponding segment from GluClα counteracted the increased expression observed upon introduction of the GluClα signal peptide. Removal of both RxR ER retention motifs enhanced expression approximately 4-fold whereas the α signal peptide chimera had approximately 8-fold enhanced expression. The α signal peptide chimera gave currents comparable to other Cys-loop receptors, and therefore mutant receptors not responding to glutamate at the applied concentrations are assumed to be nonfunctional rather than inadequately expressed. No shift in EC50 was observed for any of the chimeras or mutants examined, with the exception of a less than than 3-fold increase in EC50 of the α signal peptide chimera.
Evaluating Potential Cation−π Interactions
A cation−π interaction between agonist and receptor is a universal feature of Cys-loop receptors. These binding sites are rich in aromatic residues, and in most nAChRs and the 5HT3A receptor, a Trp on loop B provides the π system. Receptors for smaller agonists such as GABA and glycine do not typically have Trp residues at the agonist binding site, and the cation−π interaction is formed to a Phe or Tyr. Along with loop B, aromatics on loops A and C have been shown to make cation−π interactions to agonists.
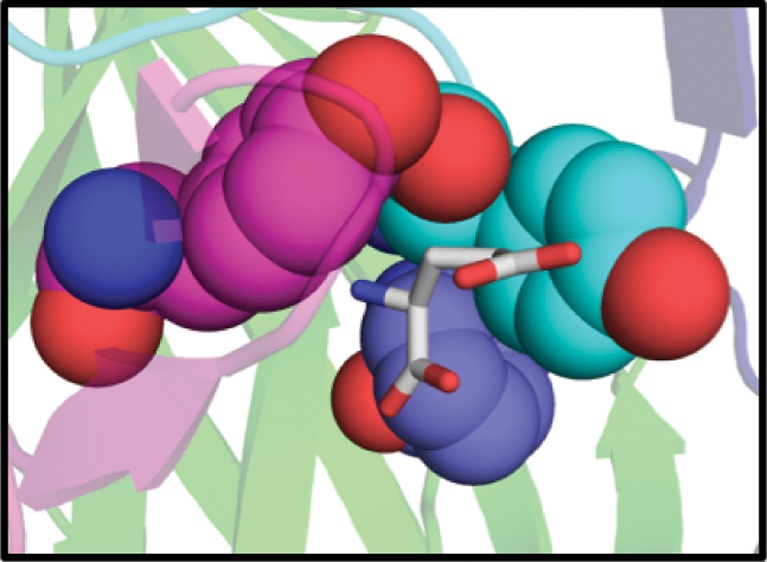
To facilitate visualization of the discussed residues in the GluClα crystal structure, the residue identifier for the corresponding GluClα amino acid is included after the GluClβ number (ex. Y206(200) corresponds to Yβ(α)). The GluClα numbering is included for the first mention of the residue only. The glutamate-bound GluClα crystal structure suggests a strong cation−π interaction between Y206(200) on the C loop and the protonated amine of glutamate (Figure 1). The crystal structure also indicates the potential for a weaker, but still significant, electrostatic interaction between the α carbon of the ligand (which carries a substantial positive charge) and the face of Y156(151), located on the B loop. Aromatic residues occur at the aligning positions of most Cys-loop receptors and have been previously shown to participate in important cation−π interactions in other receptors with their corresponding ligands.27,38−41 Additionally, a phenylalanine, F96(91), on loop A and a tryptophan on the D loop of the complementary face, W62(59), are conserved members of the aromatic binding box. However, these residues are not predicted to participate in an electrostatic interaction with the ligand or any other residue. A major difference between the GluCl receptors and other Cys-loop receptors is the absence of a second aromatic residue on the C loop (C1).
Figure 1.
Glutamate binding site and predicted electrostatic interactions between the ligand and its surrounding amino acids. Residues from the primary face are colored in green and the complementary face in blue.
We have established a general protocol for evaluating cation−π interactions,16 in which the aromatic of interest is substituted with electron withdrawing groups that are known to diminish the cation−π binding ability of the ring (Figure 2). A clear correlation between cation−π binding ability and receptor function, as reported by EC50, indicates a functionally significant cation−π interaction. An especially relevant system is a series of progressively fluorinated phenylalanine derivatives, which monotonically lower the cation−π binding ability. Such a study shows no electrostatic interaction between the ligand and either F96 or Y156, as indicated by the very small losses-of-function upon incorporation of F3–Phe, a highly electron-deficient ring system (Table 2).
Figure 2.
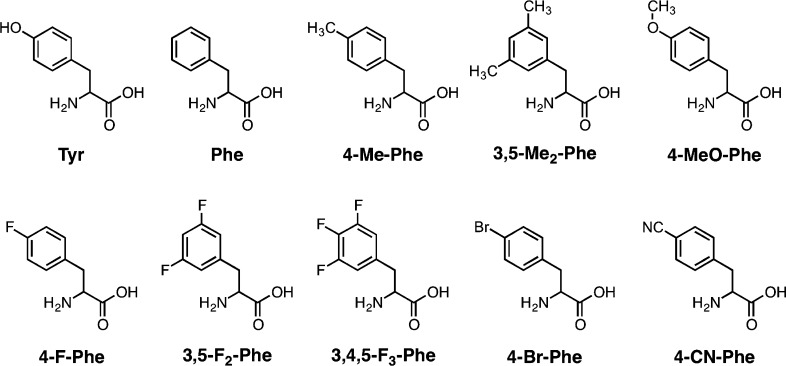
Chemical structures of amino acids used in this study.
Table 2. Unnatural Amino Acid Mutagenesis of Aromatic Binding Site Residuesa.
| EC50 (mM) | Hill | n | Imax (μA) | fold shift | |
|---|---|---|---|---|---|
| WT | 0.93 ± 0.02 | 1.8 ± 0.1 | 26 | 2–13 | |
| T6′S | 0.013 ± 0.001 | 2.7 ± 0.2 | 17 | 2–13 | |
| Y156F | 15 ± 1 | 2.3 ± 0.2 | 15 | 1–5 | |
| Y206F | 0.10 ± 0.01 | 2.6 ± 0.3 | 14 | 0.14–7 | |
| F96TAG | |||||
| Phe | 0.012 ± 0.001 | 2.8 ± 0.3 | 12 | 0.1–4 | |
| F1–Phe | 0.080 ± 0.002 | 2.8 ± 0.2 | 7 | 0.1–0.5 | 7 |
| F2–Phe | 0.0047 ± 0.0001 | 4.1 ± 0.5 | 12 | 0.04–1 | 1/3 |
| F3–Phe | 0.039 ± 0.001 | 2.4 ± 0.2 | 11 | 0.06–0.6 | 3 |
| Y156TAG | |||||
| Phe | 17 ± 1 | 1.7 ± 0.1 | 15 | 1–4 | |
| OMe–Phe | no response | ||||
| F1–Phe | 14 ± 1 | 2.0 ± 0.1 | 20 | 0.1–5 | 1/1.2 |
| F2–Phe | 49 ± 3 | 1.8 ± 0.1 | 13 | 0.2–4 | 3 |
| F3–Phe | 65 ± 5 | 1.7 ± 0.1 | 11 | 0.3–2 | 4 |
| Cha | no response | ||||
| Y206TAG | |||||
| Phe | 0.16 ± 0.01 | 3.0 ± 0.2 | 8 | 0.07–0.30 | |
| F1–Phe | 0.64 ± 0.01 | 2.8 ± 0.1 | 9 | 0.04–0.34 | 4 |
| F2–Phe | >100 | 8 | 0.1–0.6 | >250 | |
| OMe–Phe | 0.56 ± 0.01 | 1.8 ± 0.1 | 11 | 0.1–1.4 | 4 |
| Br–Phe | 0.98 ± 0.03 | 2.5 ± 0.1 | 15 | 0.08–6 | 6 |
| CN–Phe | 19 ± 1 | 2.8 ± 0.2 | 14 | 0.8–6 | 119 |
| Me–Phe | 0.098 ± 0.003 | 2.4 ± 0.1 | 12 | 0.05–7 | 1/1.8 |
Data for F96 and Y156 were obtained from a receptor containing the α signal peptide for enhanced functional expression and data for Y206 were obtained from a wild type receptor. All mutants have the T6′S background mutation.
In contrast, electron-withdrawing groups added to Y206 strongly impacted receptor function. As before, we study substituted Phe analogues to avoid complications associated with lowering the pKa of a Tyr residue when electron-withdrawing groups are added. Most telling is the large effect for 4-CN-Phe vs 4-Br-Phe. The two substituents present a similar steric perturbation, but only CN is strongly deactivating in a cation−π interaction. We were unable to study the full series of fluorinated Phe derivatives that we have previously employed, because 3,5-F2–Phe gave receptors that did not reach their maximal response upon application of our highest concentration of glutamate, 100 mM. There is apparently a strong steric penalty for substituents in the meta position at residue 206, where the side chains of T201(195) and T203(197) could be impacted, as incorporation of 3,5-Me2-Phe also resulted in nonfunctional receptors. As such, our study is restricted to 4-substituted Phe derivatives. With this more limited data set, a linear plot of calculated cation−π binding energy and the log ratio of the mutant EC50 to the wild type could be obtained (Figure 3). Although this is a limited data set, we feel the correlation of Figure 3, along with the very large CN/Br ratio (∼20) makes a compelling case for a cation−π interaction to Y206.
Figure 3.
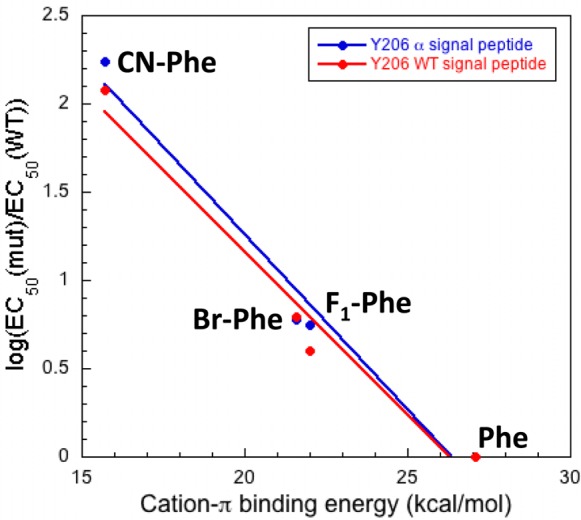
Plot of cation−π binding energy vs mutant receptor loss-of-function at Y206. A linear trend is indicative of an electrostatic interaction with the face of the residue examined. Data for both the wild type and α signal peptide-containing templates are plotted together and indicate no effect of altering the signal peptide on receptor function.
In addition to the 3,5-F2–Phe mutation, full saturation of the dose–response relation was not seen for 4-CN-Phe in the α signal peptide-containing template (SI Figure S4). The α signal peptide was required to obtain suppression data at the F96 and Y156 sites; however, suppression data could be obtained for both the wild type and α signal peptide-containing Y206 mutations (Table 2 and SI Table S3). Data were obtained in both templates for Y206 mutations to ensure that the signal peptide did not affect receptor function. To estimate an EC50 value, obtained data were fit to the Hill equation. As shown in Figure 3, very similar results are obtained for receptors with either the α or the β signal peptide, providing evidence that altering the signal peptide has no effect on receptor function once it reaches the plasma membrane, and boosting confidence in the EC50 values obtained for the receptors with the α signal peptide. The Y206 3,5-F2–Phe mutant in both constructs began to respond to glutamate but did not near a plateau. These data were fit to the Hill equation (SI Figure S4); however, we do not believe that the resulting EC50 values represent an accurate indication of receptor function. We have therefore reported this mutation as having an EC50 >100 mM. The D loop tryptophan was not examined, because it is not located near the binding site in the crystal structure.
Probing Other Noncovalent Interactions
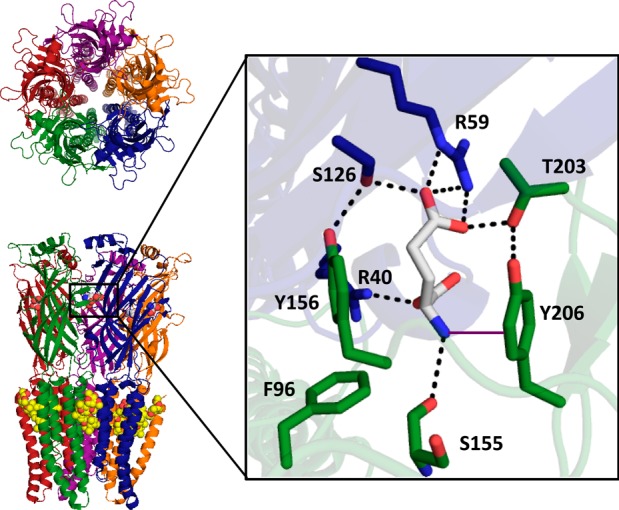
In addition to a strong cation−π interaction, other noncovalent interactions between glutamate and the receptor were identified. Two arginine residues on the complementary subunit, R40(37) and R59(56), were predicted by the crystal structure to form ionic interactions with the main chain carboxylate and the side chain carboxylate of glutamate, respectively (Figure 1). These interactions are confirmed through receptor mutagenesis. The R59 residue appears to be more sensitive to mutagenesis than R40, because R59 could not tolerate mutation to alanine, whereas R40A demonstrated a large but measurable loss-of-function of approximately 140-fold (Table 3). Mutagenesis of either residue to glutamate resulted in a nonfunctional receptor.
Table 3. Conventional Mutagenesis of Binding Site Residuesa.
| mutation | EC50 (mM) | Hill | n | fold shift | Ω | ΔΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| WT (T6′S) | 0.0080 ± 0.0005 | 2.4 ± 0.3 | 27 | |||
| R40A | 1.1 ± 0.1 | 2.1 ± 0.1 | 17 | 140 | ||
| R40E | no response | |||||
| R59A | no response | |||||
| R59E | no response | |||||
| Y206A | no response | |||||
| Y206F | 0.10 ± 0.01 | 2.6 ± 0.3 | 14 | 13 | ||
| T203A | 0.64 ± 0.01 | 2.3 ± 0.1 | 19 | 80 | ||
| T203A/Y206F | 0.21 ± 0.01 | 2.2 ± 0.1 | 18 | 26 | 1/37 | 2.1 |
| Q174A | 0.11 ± 0.01 | 2.4 ± 0.2 | 14 | 14 | ||
| N202A | 0.010 ± 0.001 | 3.3 ± 0.2 | 16 | 1.25 | ||
| Q174A/Y206F | 0.92 ± 0.04 | 2.2 ± 0.2 | 13 | 115 | 0.67 | 0.23 |
| Y156A | no response | |||||
| Y156F | 6.4 ± 0.4 | 1.6 ± 0.1 | 17 | 800 | ||
| S126A | 4.7 ± 0.1 | 2.5 ± 0.1 | 22 | 590 | ||
| S126A/Y156F | no response | |||||
All mutants have the T6′S background mutation.
Two intersubunit hydrogen bond networks are predicted by the GluClα crystal structure: one between the C loop of the primary subunit and the F loop of the complementary subunit and another between the B loop of the primary subunit and the E loop of the complementary subunit (Figure 4). Motivation to study the former comes from previous studies in the muscle-type nAChR, where an intersubunit hydrogen bond between an aspartate side chain (γD174/δD180) on the complementary subunit and a backbone nitrogen (αS191) on the β-hairpin turn of the C loop was shown to be important for channel gating.42 This backbone nitrogen was later shown to be optimally positioned by a hydrogen bond network initiating from a neighboring vicinal disulfide bond (αC192/αC193) on the C loop.43 While the GluCl receptors, as well as all non-α nAChRs, lack this vicinal disulfide, other structural motifs may have evolved to perform the same function.
Figure 4.
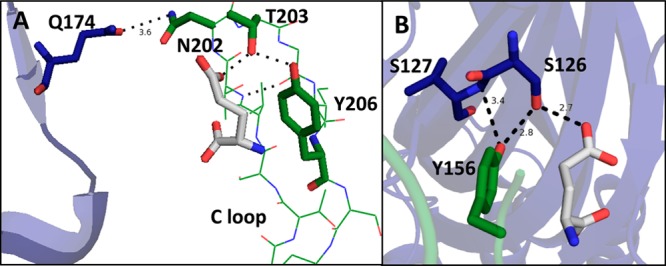
Predicted hydrogen bond interactions between the primary and complementary subunits near the ligand-binding site in the 3RIF crystal structure. Residues on the primary face are shown in green, residues on the complementary face in blue, and glutamate in white. Hydrogen bond network between (A) the C loop of the primary subunit and the F loop of the complementary subunit and (B) the B loop of the primary subunit and the E loop of the complementary subunit.
The GluClα crystal structure predicts an intrasubunit hydrogen bond between the side chains of T203(197) and Y206(200) on loop C in approximately the same relative position as the vicinal disulfide. Additionally, a glutamine, Q174(169), is located two residues away from the aspartates of the F loop in the γ and δ subunits of the nAChR and is 3.6 Å from the side chain of an asparagine on the β-hairpin turn of the C loop, N202(196), suggesting the possibility of a hydrogen bond. While these interactions are different than those observed in the muscle type nAChR, they have the potential to be functionally analogous.
To analyze these potential interactions, mutant cycle analysis was performed. The data indicate a strong functional coupling between the side chains of T203 and Y206, with a coupling strength of 2.1 kcal/mol (Table 3), between the T203A and Y206F mutants. A 14-fold loss-of-function was observed for the Q174A mutant, providing evidence that this side chain is important for receptor function. Interestingly, the N202A mutant gives receptors with a wild type EC50, despite an indication of a possible hydrogen bond between Q174 and N202 in the crystal structure. No long-range functional coupling was observed between Y206 and Q174. These findings do not support a long-range functional coupling between structural and/or ligand-binding elements on the C loop and an intersubunit hydrogen bond with the F loop, although such an interaction is seen in the nAChR. The only other polar neighbor of Q174 is an asparagine also on the complementary subunit, N36(33), suggesting that no functionally important intersubunit interaction exists between the C loop of the primary subunit and the F loop of the complementary subunit. It is possible that the salt-bridges formed between the two arginine residues on the complementary subunit and the carboxylates of the ligand are sufficient to shape the ligand binding site on the complementary side, making an additional interaction between the C and F loops redundant.
While the functional coupling between the side chains of T203 and Y206 is likely unrelated to the effects caused by the vicinal disulfide, it provides support for the ligand-binding interactions previously discussed and predicted by the crystal structure. T203 is predicted to act as a hydrogen bond donor to the side chain carboxylate of glutamate and as a hydrogen bond acceptor to the hydroxyl of Y206. The hydrogen bond between Y206 and T203 likely strengthens the cation−π binding energy of the tyrosine due to an increase in electrostatic potential on the ring system, caused by the electron donating properties of the acidic phenol. These effects were previously predicted with gas phase calculations of phenol and formamide, a hydrogen bond acceptor, where a hydrogen bond significantly enhanced a cation−π interaction in the gas phase.44 This may account for why tyrosine, a weaker cation−π binder than tryptophan in isolation, is capable of forming such a strong cation−π interaction with glutamate in this receptor and why tyrosines are overrepresented in cation−π interactions in Cys-loop receptors relative to phenylalanine. It can also be argued that the hydrogen bond exists to position Y206 in an orientation optimal for a strong cation−π interaction. It is important to note that the cation−π interaction determined here was measured with phenylalanine derivatives that lack the hydrogen bond to T203. Data indicate a cation−π binding strength similar to other primary amines as indicated by the slope of the cation−π plot.38,40,41,45 Regardless of mechanism, we experimentally observe the importance of this hydrogen bond with the Y206F mutation, which results in a 13-fold loss of function. The directionality of hydrogen bonding predicted by the crystal structure is confirmed by incorporation of 4-MeO-Phe at Y206. This mutation does not reverse the loss-of-function seen in the Y206F mutant and therefore provides evidence that Y206 is acting as a hydrogen bond donor. The T203A mutation resulted in an 80-fold loss-of-function, likely caused by breaking its interaction to both glutamate and Y206.
In addition to intersubunit interactions between the C and F loops, the crystal structure suggests a hydrogen bond interaction between the side chains of Y156 on the B loop of the primary subunit and S126 on the E loop of the complementary subunit. Mutagenesis of these residues individually to remove the polar side chain resulted in a large EC50 increase of 800-fold and 590-fold, respectively (Table 3). These large and similar losses-of-function indicate that an interaction between the two side chains is likely to occur and is important for receptor function; however, functional coupling could not be determined because the EC50 of the double mutant could not be measured. The high EC50 values of the single mutants are near the limit of our detection, and therefore, any additional loss-of-function would escape detection.
In other anion-selective Cys-loop receptors where a tyrosine is found at the aromatic position on loop B, different effects are observed upon mutagenesis of the tyrosine to phenylalanine. In the glycine receptor, this residue is naturally a phenylalanine, and therefore, a hydrogen bond with the complementary subunit is not possible. In the GABAC receptor, a less than 5-fold loss of function is observed upon mutagenesis of tyrosine to phenylalanine.40 However, in the GABAA receptor, a large 400-fold loss-of-function was observed, similar to that seen here in the GluClβ receptor.45 Interestingly, substitution of tyrosine with 4-MeO-Phe was able to recover the receptor to wild type EC50 in GABAA, indicating that the tyrosine acts as a hydrogen bond acceptor, whereas in GluClβ, substitution with 4-MeO-Phe led to a nonfunctional receptor indicating that it acts as a donor. These data suggest that the interactions between the primary and complementary subunits vary, even between highly related receptors, and that a hydrogen bond at this site is not a universal feature of Cys-loop receptors.
In summary, a collection of interactions that were predicted by the crystal structure of the homologous GluClα receptor was examined in the GluClβ receptor. Unnatural amino acid mutagenesis indicates a strong cation−π interaction with a tyrosine residue on loop C, Y206. This ligand-binding interaction is a conserved feature of the mammalian Cys-loop receptors studied to-date and is found here for the second time in a distantly related nematode Cys-loop receptor.46 While this interaction shows that many of the features between distant classes of Cys-loop receptors are conserved, many interactions were found that are receptor-specific. An inter-subunit hydrogen bond between loops C and F shown to be important in the muscle type nAChR is absent in GluClβ. Conversely, an important inter-subunit interaction between loops B and E in GluClβ is absent in many more closely related anion-selective homologues. These data indicate that while the crystal structure of a receptor can serve as an excellent starting point for other homologous receptors, additional information can be obtained from functional studies coupled with chemical biology.
Methods
Molecular Biology
The mammalian codon optimized GluClβ receptor from C. elegans(18) was subcloned into the pGEMhe vector and the stop codon of the receptor was mutated from TAG to TGA. Mutagenesis of the GluClβ receptor was performed using the QuikChange protocol (Stratagene), and for nonsense suppression experiments, a TAG codon was mutated into the site of interest. The cDNA was linearized using SbfI (New England Biolabs) and mRNA was produced from the linearized plasmids by using the T7 mMessage Machine kit (Ambion).
74mer THG73 tRNA was in vitro transcribed from a DNA oligonucleotide template containing two 5′ methoxy (C2′ position) nucleotides to site-specifically truncate transcription by the T7 MEGAshortscript kit (Ambion).19 Amino acids chemically appended to dCA were ligated to the 74mer tRNA, resulting in full-length acylated tRNA, using methods previously described.20 Acylation of tRNA was confirmed using MALDI mass spectrometry using a 3-hydroxypicolinic acid matrix. The NVOC protecting group on the amino acid was removed immediately prior to injection through a 5 min irradiation step using a 1 kW xenon lamp with WG-335 and UG-11 filters.
Chimeras
Chimeras were synthesized by mutating in an EcoRI restriction enzyme site (GAATTC) in both the GluClα and GluClβ receptors at the desired cut site. First, the inherent EcoRI site in the GluClα M3-M4 loop (ATGAATTCC) was removed using silent mutations (ATGAACAGC) to eliminate undesired cutting. The mutated receptors were then cut with EcoRI and HindIII (5′ of the encoded gene) and the fragments separated by gel electrophoresis on a 1% agarose gel. The resulting bands were isolated and purified using a gel extraction kit (Qiagen) and the desired GluClα and GluClβ fragments were ligated together using DNA ligase. The engineered EcoRI restriction site was then mutated back to the wild type α/β or β/α sequence and mRNA was produced as described above.
Oocyte Preparation and Injection
Stage V–VI Xenopus laevis oocytes were harvested and injected with RNA as previously described.20 For nonsense suppression experiments, 37.5 ng of receptor mRNA was coinjected with excess deprotected ∼1 μg/μL tRNA solution both 48 and 24 h before recording. For conventional mutagenesis, 12.5 ng of receptor mRNA was injected 48 h before recording. As a negative control, all nonsense suppression sites were tested with a full length 76mer tRNA lacking an attached amino acid to ensure no read-through expression was observed.
Data Collection and Analysis
All oocyte experiments were performed on an OpusXpress 6000A (Axon Instruments) using two-electrode voltage clamp mode. The recording buffer was calcium-free ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES, pH 7.5). The initial holding potential was −60 mV. Data were sampled at 125 Hz and filtered at 50 Hz. A calcium-free ND96 prewash was applied for 30 s (1 mL/min) followed by application of agonist for 15 s (4 mL/min). Agonist was then washed out with calcium-free ND96 buffer for 116 s (3 mL/min). Agonist-induced currents were measured from the baseline. Glutamate dose solutions were made in calcium-free ND96 buffer from a 50 mM (pH 7.5) stock solution. The maximum dose of 100 mM glutamate was made from the glutamate-HCl salt in water and brought to a pH of 7.5.
Data were fit to the Hill equation, Inorm = 1/(1 + (EC50/A))nH, where Inorm is the normalized current peak at [agonist] = A, EC50 is the concentration of agonist that elicits a half-maximum response, and nH is the Hill coefficient. EC50 values were obtained by averaging the Inorm values for each agonist concentration and fitting those values to the Hill equation. ΔΔG values were calculated by the equation ΔΔG = −RT ln Ω, where Ω = [EC50(mut1,2) × EC50(WT)]/[EC50(mut1) × EC50(mut2)].
Acknowledgments
This work was supported by the National Institutes of Health grant NS034407. We are also grateful for support from the Beckman Institute Zebrafish/Xenopus/Lamprey Facility.
Supporting Information Available
Additional tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Thompson A. J.; Lester H. A.; Lummis S. C. (2010) The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43, 449–499. [DOI] [PubMed] [Google Scholar]
- Coe J. W.; Brooks P. R.; Vetelino M. G.; Wirtz M. C.; Arnold E. P.; Huang J.; Sands S. B.; Davis T. I.; Lebel L. A.; Fox C. B.; Shrikhande A.; Heym J. H.; Schaeffer E.; Rollema H.; Lu Y.; Mansbach R. S.; Chambers L. K.; Rovetti C. C.; Schulz D. W.; Tingley F. D. 3rd; O’Neill B. T. (2005) Varenicline: An α4β2 nicotinic receptor partial agonist for smoking cessation. J. Med. Chem. 48, 3474–3477. [DOI] [PubMed] [Google Scholar]
- Cupp E. W.; Bernardo M. J.; Kiszewski A. E.; Collins R. C.; Taylor H. R.; Aziz M. A.; Greene B. M. (1986) The effects of ivermectin on transmission of Onchocerca volvulus. Science 231, 740–742. [DOI] [PubMed] [Google Scholar]
- Miyazawa A.; Fujiyoshi Y.; Stowell M.; Unwin N. (1999) Nicotinic acetylcholine receptor at 4.6 A resolution: Transverse tunnels in the channel wall. J. Mol. Biol. 288, 765–786. [DOI] [PubMed] [Google Scholar]
- Unwin N. (2005) Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 346, 967–689. [DOI] [PubMed] [Google Scholar]
- Brejc K.; van Dijk W. J.; Klaassen R. V.; Schuurmans M.; van Der Oost J.; Smit A. B.; Sixma T. K. (2001) Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276. [DOI] [PubMed] [Google Scholar]
- Hilf R. J.; Dutzler R. (2008) X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379. [DOI] [PubMed] [Google Scholar]
- Hilf R. J.; Dutzler R. (2009) Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118. [DOI] [PubMed] [Google Scholar]
- Bocquet N.; Nury H.; Baaden M.; Le Poupon C.; Changeux J. P.; Delarue M.; Corringer P. J. (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114. [DOI] [PubMed] [Google Scholar]
- Hibbs R. E.; Gouaux E. (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W. C. (1985) Ivermectin: An update. Parasitol Today 1, 10–16. [DOI] [PubMed] [Google Scholar]
- Campbell W. C.; Fisher M. H.; Stapley E. O.; Albers-Schonberg G.; Jacob T. A. (1983) Ivermectin: A potent new antiparasitic agent. Science 221, 823–828. [DOI] [PubMed] [Google Scholar]
- Aziz M. A.; Diallo S.; Diop I. M.; Lariviere M.; Porta M. (1982) Efficacy and tolerance of ivermectin in human onchocerciasis. Lancet 2, 171–173. [DOI] [PubMed] [Google Scholar]
- Lester H. A.; Dibas M. I.; Dahan D. S.; Leite J. F.; Dougherty D. A. (2004) Cys-loop receptors: New twists and turns. Trends Neurosci. 27, 329–336. [DOI] [PubMed] [Google Scholar]
- Miller P. S.; Smart T. G. (2010) Binding, activation, and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 31, 161–174. [DOI] [PubMed] [Google Scholar]
- Van Arnam E. B.; Dougherty D. A. (2014) Functional probes of drug–receptor interactions implicated by structural studies: Cys-loop receptors provide a fertile testing ground. J. Med. Chem. 10.1021/jm500023m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter A.; Cully D. F.; Schaeffer J. M.; Liu K. K.; Arena J. P. (1996) An amino acid substitution in the pore region of a glutamate-gated chloride channel enables the coupling of ligand binding to channel gating. J. Biol. Chem. 271, 16035–16039. [DOI] [PubMed] [Google Scholar]
- Slimko E. M.; Lester H. A. (2003) Codon optimization of Caenorhabditis elegans GluCl ion channel genes for mammalian cells dramatically improves expression levels. J. Neurosci. Methods 124, 75–81. [DOI] [PubMed] [Google Scholar]
- Kao C.; Zheng M.; Rudisser S. (1999) A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 5, 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M. W.; Gallivan J. P.; Silverman S. K.; Labarca C. G.; Dougherty D. A.; Lester H. A. (1998) In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 293, 504–529. [DOI] [PubMed] [Google Scholar]
- Pertzoff V. A. (1933) The solubility of glutamic acid in water and certain organic solvents. J. Biol. Chem. 100, 97–104. [Google Scholar]
- Filatov G. N.; White M. M. (1995) The role of conserved leucines in the M2 domain of the acetylcholine receptor in channel gating. Mol. Pharmacol. 48, 379–384. [PubMed] [Google Scholar]
- Labarca C.; Nowak M. W.; Zhang H.; Tang L.; Deshpande P.; Lester H. A. (1995) Channel gating governed symmetrically by conserved leucine residues in the M2 domain of nicotinic receptors. Nature 376, 514–516. [DOI] [PubMed] [Google Scholar]
- Frazier S. J.; Cohen B. N.; Lester H. A. (2013) An engineered glutamate-gated chloride (GluCl) channel for sensitive, consistent neuronal silencing by ivermectin. J. Biol. Chem. 288, 21029–21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek A. N.; Grassi F.; Meyer E. M.; Papke R. L. (2005) An α7 nicotinic acetylcholine receptor gain-of-function mutant that retains pharmacological fidelity. Mol. Pharmacol. 68, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Yakel J. L.; Lagrutta A.; Adelman J. P.; North R. A. (1993) Single amino acid substitution affects desensitization of the 5-hydroxytryptamine type 3 receptor expressed in Xenopus oocytes. Proc. Natl. Acad. Sci. U.S.A. 90, 5030–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu X.; Puskar N. L.; Shanata J. A.; Lester H. A.; Dougherty D. A. (2009) Nicotine binding to brain receptors requires a strong cation−π interaction. Nature 458, 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta C. B.; Dilworth C. N.; Lester H. A.; Dougherty D. A. (2014) Probing the noncanonical interface for agonist interaction with an α5 containing nicotinic acetylcholine receptor. Neuropharmacology 77, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W. G.; Gallivan J. P.; Zhang Y. O.; Li L. T.; Lester H. A.; Dougherty D. A. (1998) From ab initio quantum mechanics to molecular neurobiology: A cation−π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A. 95, 12088–12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney P. C.; Zhang H.; Zhong W.; Dougherty D. A.; Lester H. A. (1996) Determinants of nicotinic receptor gating in natural and unnatural side chain structures at the M2 9′ position. Neuron 17, 1221–1229. [DOI] [PubMed] [Google Scholar]
- Zerangue N.; Schwappach B.; Jan Y. N.; Jan L. Y. (1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22, 537–548. [DOI] [PubMed] [Google Scholar]
- Boyd G. W.; Doward A. I.; Kirkness E. F.; Millar N. S.; Connolly C. N. (2003) Cell surface expression of 5-hydroxytryptamine type 3 receptors is controlled by an endoplasmic reticulum retention signal. J. Biol. Chem. 278, 27681–27687. [DOI] [PubMed] [Google Scholar]
- Mossessova E.; Bickford L. C.; Goldberg J. (2003) SNARE selectivity of the COPII coat. Cell 114, 483–495. [DOI] [PubMed] [Google Scholar]
- Xu J.; Zhu Y.; Heinemann S. F. (2006) Identification of sequence motifs that target neuronal nicotinic receptors to dendrites and axons. J. Neurosci. 26, 9780–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracun S.; Harkness P. C.; Gibb A. J.; Millar N. S. (2008) Influence of the M3-M4 intracellular domain upon nicotinic acetylcholine receptor assembly, targeting, and function. Br. J. Pharmacol. 153, 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M.; Mulet J.; Aldea M.; Gerber S.; Sala S.; Sala F.; Criado M. (2009) Role of the N-terminal α-helix in biogenesis of α7 nicotinic receptors. J. Neurochem 108, 1399–1409. [DOI] [PubMed] [Google Scholar]
- Pons S.; Sallette J.; Bourgeois J. P.; Taly A.; Changeux J. P.; Devillers-Thiery A. (2004) Critical role of the C-terminal segment in the maturation and export to the cell surface of the homopentameric α7–5HT3A receptor. Eur. J. Neurosci 20, 2022–2030. [DOI] [PubMed] [Google Scholar]
- Beene D. L.; Brandt G. S.; Zhong W.; Zacharias N. M.; Lester H. A.; Dougherty D. A. (2002) Cation−π interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: The anomalous binding properties of nicotine. Biochemistry 41, 10262–10269. [DOI] [PubMed] [Google Scholar]
- Puskar N. L.; Xiu X. A.; Lester H. A.; Dougherty D. A. (2011) Two neuronal nicotinic acetylcholine receptors, α4, β4, and α7, show differential agonist binding modes. J. Biol. Chem. 286, 14618–14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis S. C.; D L. B.; Harrison N. J.; Lester H. A.; Dougherty D. A. (2005) A cation−π binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem. Biol. 12, 993–997. [DOI] [PubMed] [Google Scholar]
- Pless S. A.; Millen K. S.; Hanek A. P.; Lynch J. W.; Lester H. A.; Lummis S. C.; Dougherty D. A. (2008) A cation−π interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J. Neurosci. 28, 10937–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleitsman K. R.; Kedrowski S. M. A.; Lester H. A.; Dougherty D. A. (2008) An intersubunit hydrogen bond in the nicotinic acetylcholine receptor that contributes to channel gating. J. Biol. Chem. 283, 35638–35643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. P.; Gleitsman K. R.; Lester H. A.; Dougherty D. A. (2011) Evidence for an extended hydrogen bond network in the binding site of the nicotinic receptor: Role of the vicinal disulfide of the α1 subunit. J. Biol. Chem. 286, 32251–32258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecozzi S.; West A. P.; Dougherty D. A. (1996) Cation−π interactions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc. Natl. Acad. Sci. U.S.A. 93, 10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett C. L.; Hanek A. P.; Lester H. A.; Dougherty D. A.; Lummis S. C. (2007) Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation−π interaction between GABA and β 2Tyr97. J. Neurosci. 27, 886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu T. W.; Lester H. A.; Dougherty D. A. (2003) Different binding orientations for the same agonist at homologous receptors: A lock and key or a simple wedge?. J. Am. Chem. Soc. 125, 6850–6851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.