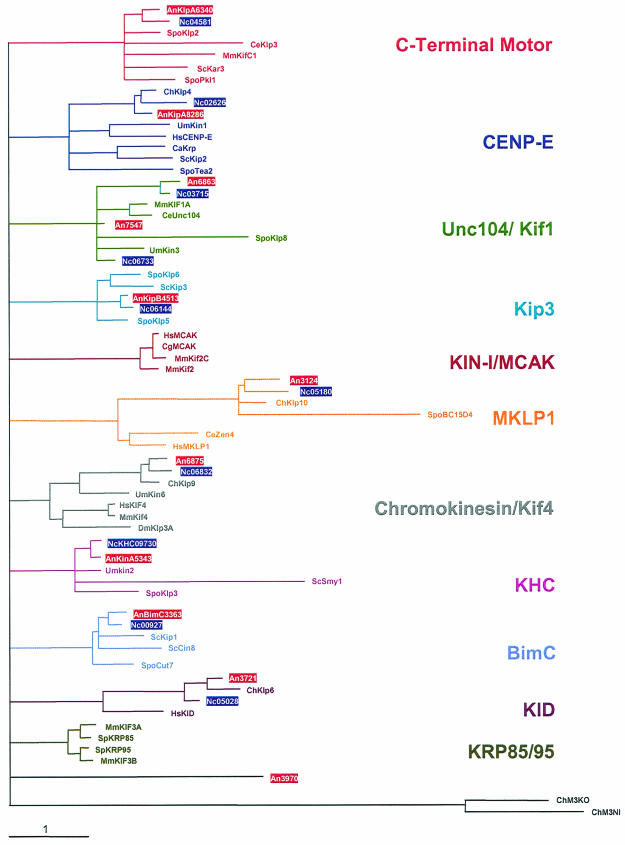
FIG. 1.
Relatedness analysis of the 11 A. nidulans and 10 N. crassa kinesins. A most likely phylogenetic tree for 74 kinesins was built with Treepuzzle (http://www.tree-puzzle.de/), using a maximum-likelihood algorithm. For an evaluation of the statistical significance of the topology, 25,000 replicating puzzle steps were performed. The substitution model Whelan-Goldman 2000 was used because it produced a consensus tree which was in good agreement with the published data (37). For the construction of the tree, we chose fungal kinesin sequences and additional kinesins from other organisms that are characteristic for the different families. Note that the A. nidulans exon-intron borders were only experimentally determined for BimC, KlpA, KinA, KipA, and KipB. In the case of N. crassa, only the conventional kinesin NcKHC has been analyzed experimentally. The other primary structures of the proteins of A. nidulans and N. crassa are based on the predictions in the annotation process at the Whitehead Institute database. The N. crassa Kip3 homologue was annotated manually. The origins of the kinesin sequences were as follows: An, A. nidulans; Nc, N. crassa; Spo, Schizosaccharomyces pombe; Ce, Caenorhabditis elegans; Mm, Mus musculus; Um, Ustilago maydis; Sc, S. cerevisiae; Ch, Cochliobolus heterostrophus; Hs, Homo sapiens; Ca, Candida albicans; Dm, Drosophila melanogaster; Cg, Cricetulus griseus; Sp, Strongylocentrotus purpuratus. The A. nidulans and N. crassa sequences are boxed.