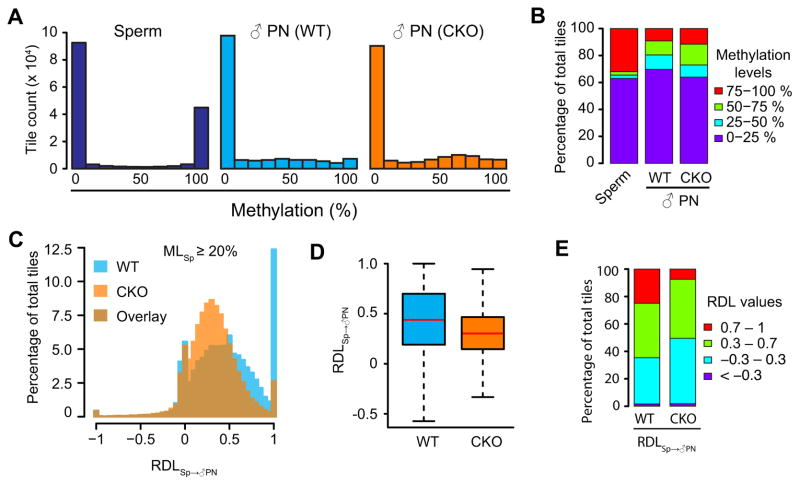
Figure 2. Loss of Tet3 partially impairs paternal DNA demethylation.
(A) Histograms of DNA methylation values across 100-bp tiles in sperm, WT and CKO paternal pronuclei. CpG sites that are at least 10x covered in all samples were examined. Note that common 10x CpG sites are more enriched in CpG-rich sequences and thus show lower overall DNA methylation level compared to that of all detected sites.
(B) Distributions of DNA methylation levels in sperm, WT, and CKO paternal pronuclei. 100-bp tiles of genomic DNA were divided into four groups based on their methylation levels as low (0–25%, purple), medium-low (25–50%, blue), medium-high (50–75%, green), and high (75–100%, red) methylation.
(C) Histograms showing the distributions of RDLSp→♂PN values of 100-bp tiles in WT and CKO paternal DNA. Only tiles that are methylated in sperm (MLSp ≥20%, n=58,101) were examined. RDLSp→♂PN is defined as [(MLSp – ML♂PN)/MLSp]. ML, DNA methylation level. RDL values less than −1 were set to −1.
(D) Boxplot of RDLSp→♂PN values. Red line represents the median. Boxes and whiskers represent for the 25th and 75th, and 2.5th and 97.5th percentiles, respectively.
(E) Distribution of RDLSp→♂PN values in WT and CKO paternal DNA. The RDL values are separated into four groups marked by red (0.7–1), green (0.3–0.7), blue (−0.3–0.3), and purple (<−0.3).