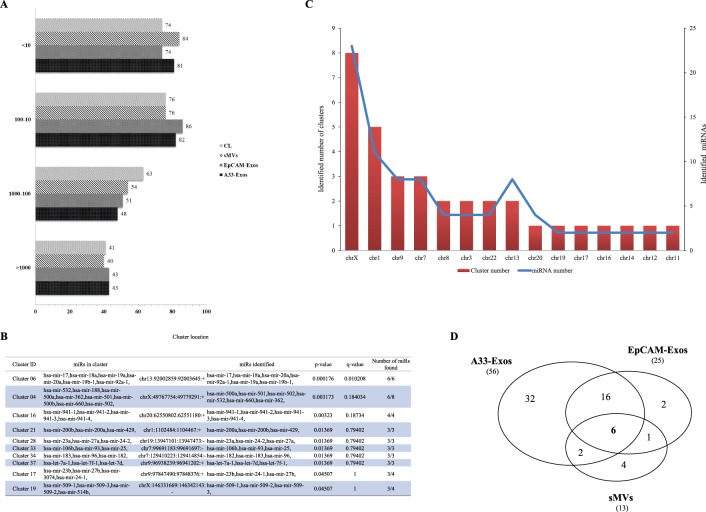
Figure 2. Characterisation of 254 highly-represented miRNAs in all four miRNA libraries (A33-Exos, EpCAM-Exos, sMVs and CL).
(A) Distribution of known miRNA sequences in CL, sMVs, A33- and EpCAM-Exos based on normalised expression values (transcripts per million reads, TPM). (B) Top 10 miRNA clusters based on analysis of the 254 miRNAs identified with respect to their location on the human reference genome (hg19). (C) Distribution of clustered miRNAs on different human chromosomes. Clustered miRNAs (including miRNA number) were identified according to their chromosomal locations, which should be within 10 k bp on the chromosome; miRNAs from the same precursor (-5p, -3p) were only considered once. (D) Three-way Venn diagram depicting the 63 miRNAs enriched in sMVs, A33- and EpCAM-Exos, relative to CL. 6 miRNAs are common to all EVs, while 22 miRNAs are common to both exosomal datasets. miRNAS selectively enriched in each EV miRNA dataset: A33-Exos 32/56, EpCAM-Exos 2/25, and sMVs 4/13.