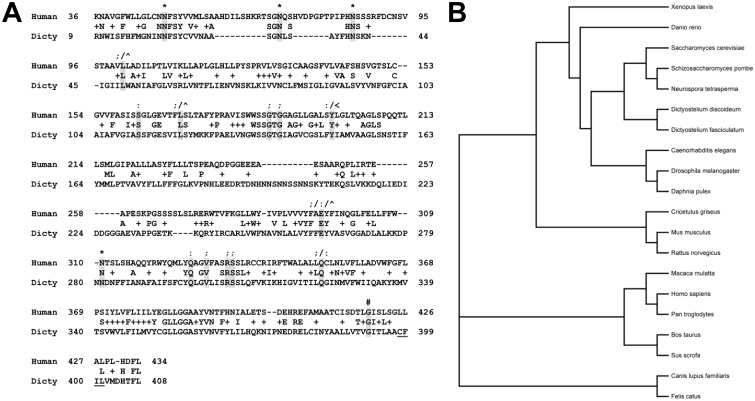
Figure 1. Bioinformatic analysis of Dictyostelium Cln3.
(A) Alignment of human CLN3 and the Dictyostelium ortholog. The following residues are conserved; N-linked glycosylation sites (*), sites of missense point mutations (;), sites of nonsense point mutations (:), target for myristoylation (#), sites that when mutated cause a slower disease progression in compound heterozygosity with the common 1.02 kb deletion mutation (∧), sites that when mutated cause a slower disease progression in homozygosity (<) [10], [13], [85]–[87]. Dictyostelium Cln3 also contains a putative prenylation motif (i.e., CFIL; underlined). (B) Phylogenetic tree showing the relationship of Dictyostelium Cln3 to CLN3 orthologs from 20 different organisms (i.e., mammals and NIH model systems).