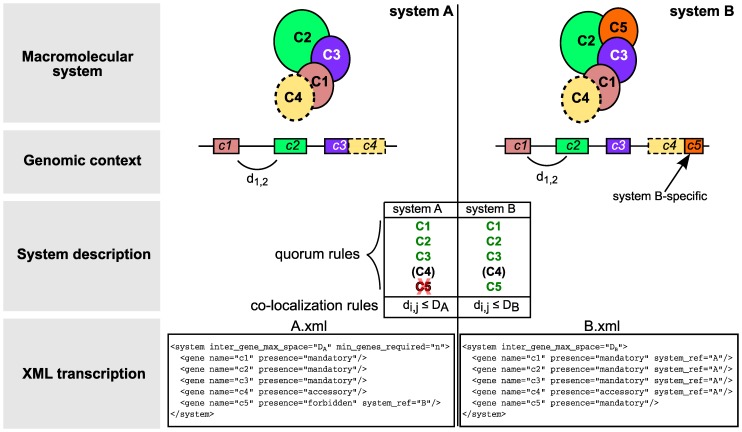
Figure 1. Modelling systems with MacSyFinder.
The components of a system assemble into macromolecular systems or correspond to a biological pathway. They are typically encoded in genomes in one or a few different loci (“Genomic context”). We illustrate how systems can be modelled and distinguished with two imaginary systems “A” and “B” that have four homologous components (C1–C4, similar colours for the two systems). The system “B” has one component that is not found in “A”(C5). The parameter inter_gene_max_space (D) defines the maximal number of genes between two consecutive components (di,j). The two systems are defined by a set of mandatory (green), accessory (black) and forbidden (red) components. The quorum rules allow relaxing the definition of the system without altering the list of its components (min_genes_required and min_mandatory_genes_required parameters in XML files). If they are not specified, a default value is computed from the number of components described in the XML files. The bottom part of the figure shows the description of the systems in the XML grammar (see the documentation in File S1). Components listed here refer to protein profiles (Fig. 3). When a component is found in several systems, it is defined only once, and can be reused in another system with the system_ref keyword. Much more complex features can be defined, including exchangeable genes, distant genes and component-specific parameters (File S1).