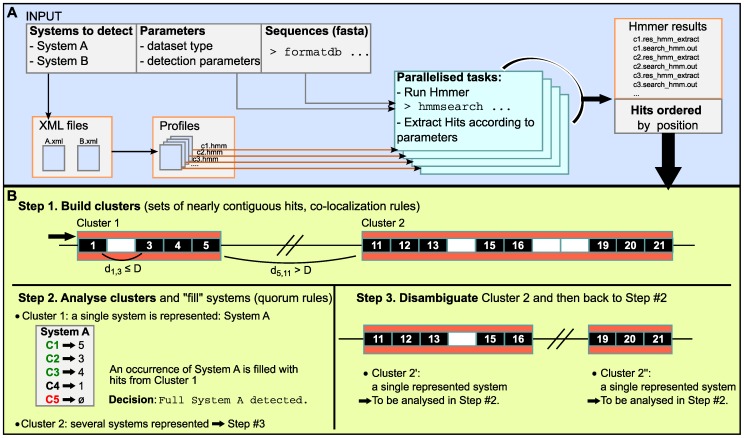
Figure 3. Functioning of MacSyFinder.
A. The user launches MacSyFinder to detect macromolecular systems A and B (example of Fig. 1). System-specific parameters are read from the corresponding XML definition files. This includes the list of the components of the systems and the corresponding HMM profiles. Other detection parameters are picked by order of priority: on the command-line, in the configuration file, and in the XML files. Sequences are indexed with the “formatdb” or “makeblastdb” tools for similarity search with the Hmmer program. MacSyFinder runs (optionally in parallel) the Hmmer searches on a non-redundant list of components' profiles. If the sequence dataset is “unordered” MacSyFinder only outputs the hits and the components detected for each type of system. B. Step #1: the co-localization criterion can be used in the ordered datasets. It involves clustering the hits separated by less than D protein-coding genes. The components described as “loner” in the XML definition files can be at any distance from other components. Step #2: the components of each cluster are used to fill the occurrences of the systems. Depending on the quorum, a cluster can describe a “full” system, or a “scattered” system. Step #3: clusters with components belonging to more than one system are split in unique systems and then re-directed separately to step #2.