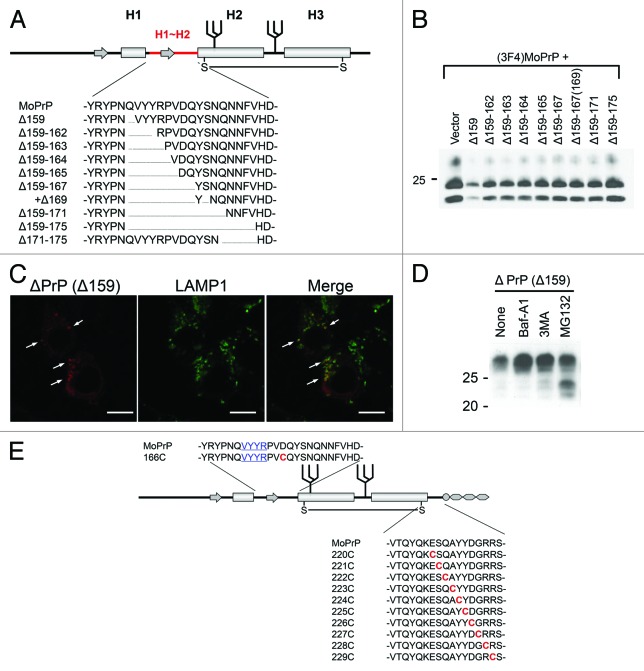
Figure 2. (A) Example of systematically-designed mutant PrPs with deletions in H1~H2, used to test in DNI assay whether the H1~H2 region is an interaction interface. (B) Representative immunoblot demonstrating inverse correlation between DNI efficiency and size of deletion in H1~H2. A conversion-competent and epitope-tagged wild-type PrP, (3F4)MoPrP, and a conversion-incompetent PrP mutant were co-transfected into 22L prion-infected N2a cells and PK-resistant PrP was detected. (C) Confocal image showing co-localization of PrPΔ159 and LAMP1. N2a cells transfected with PrPΔ159 were fixed, treated with 6M guanidine hydrochloride to remove excessive PrPΔ159 signal localized in ER, labeled with antibodies against PrP and LAMP1, and analyzed for co-localization. (D) Immunoblot showing that degradation of PrPΔ159 is inhibited by treatment with bafilomycin A1 (Baf-A1), autophagy inhibitor 3MA, or proteasome inhibitor MG132. (E) A schematic illustration of a series of mutant PrPs with two cysteine substitutions.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.