Figure 5.
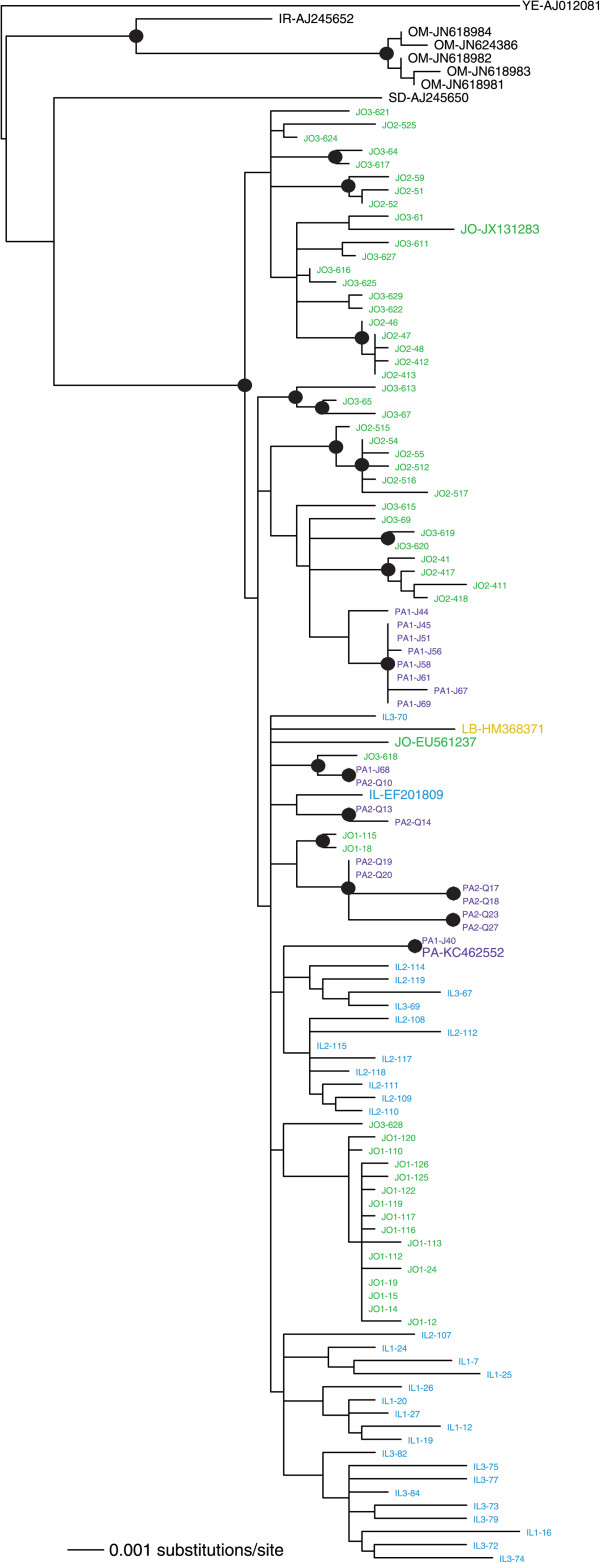
Maximum likelihood midpoint-rooted phylogeny of WmCSV DNA-A both isolated in this study and from GenBank, created in PAUP* using a Kimura two-parameter nucleotide substitution model with a gamma distribution of site heterogeneity and a parameter for the proportion of invariant sites. The locations from which various GenBank sequences were isolated, where available, were obtained from the GenBank file or from the associated publications. Sequences from Israel (IL) are shown in blue, Jordan (JO) in green, Lebanon (LB) in yellow and the regions of Palestine governed by the Palestinian Authority (PA) in purple. Sequences from all other countries are shown in black, with their two-letter country code preceding the accession number. Bootstrap support of ≥85% from both PAUP* and RaxML is shown by a solid circle.
