Fig. 2.
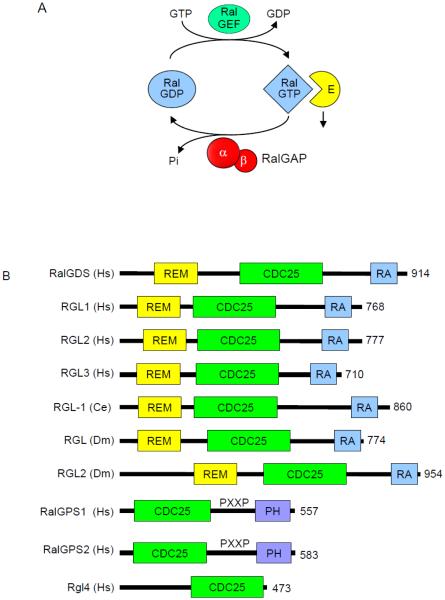
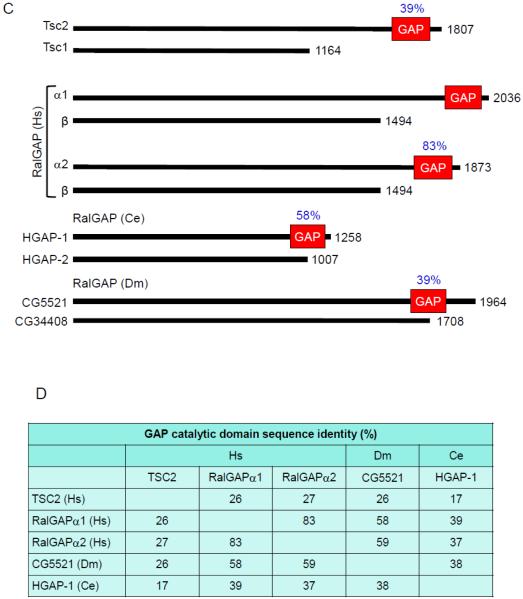
Regulators of the Ral GDP-GTP cycle. A. Regulation of Ral GDP-GTP cycling. Ral-selective GEFs and GAPs accelerate the low intrinsic exchange and GTP hydrolysis activities to promote formation of active GTP-bound and inactive GDP-bound Ral. B. The RalGEFs are highly conserved across species. All RalGEFs contain a CDC25 homology domain, which is responsible for catalytic activity. There are four human isoforms of RalGEF that contain Ras association (RA) domain. These isoforms also contain a Ras exchanger motif (REM) that likely stabilizes the CDC25 homology domain and is essential for RalGEF catalytic activity. There is one homolog in C. elegans and two in Drosophila. The RalGEF homolog in C. elegans is most similar to RalGDS. The RalGPS RalGEFs lack a REM domain and do not associate with Ras, but instead contain a pleckstrin homology (PH) domain. RGL4 contains a CDC25 homology domain, but lacks a REM, RA or PH domain. C. The RalGAPs are heterodimeric complexes formed by either a RalGAPα1 or RalGAPα2 catalytic subunit with the regulatory RalGAPβ subunit. The RalGAPβ subunit serves to regulate the catalytic activity of the RalGAPα subunits, similar to TSC1 regulation of TSC2. Percentages indicate sequence identity with the RalGAPα1 catalytic domain. Orthologs of the human RalGAPα and RalGAPβ subunits are present in C. elegans and Drosophila. Multiple sequence alignment and sequence identity was determined by ClustalW analyses and domain topology by SMART analyses.