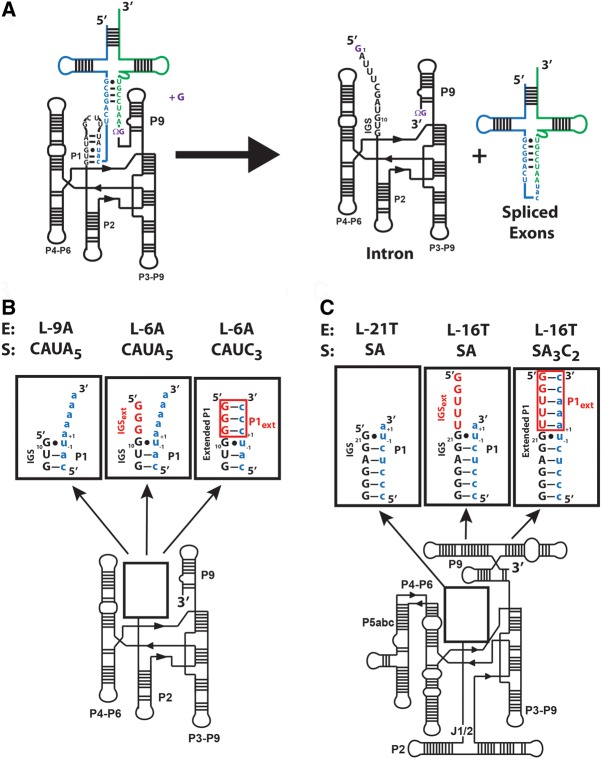
FIGURE 1.
Comparison of the ribozymes used in this work. (A) The secondary structure of the Azoarcus group I intron and its self-splicing reaction. The exogenous guanosine nucleophile, ΩG (which specifies the 3′-splice junction), and the guanosine binding site are all shown in purple. The 5′ exon is shown in blue, the 3′ exon is shown in green, and the intron is shown in black. (B) The secondary structure for the ribozymes used in this study. The numbers −1 and +1 define the nucleotides immediately 5′ and 3′ of the cleavage site, respectively. (Left) The L-9A ribozyme. Formation of base pairs with the IGS results in the formation of the P1 duplex. (Center and right) The L-6A ribozyme, containing a three nucleotide 5′ extension (5′GGG3′). The additional sequence present in the L-6A ribozyme extends the IGS and is termed the IGS extension (IGSext) and is shown in red. The IGS and the IGS extension together are referred to as the extended IGS. (Right) Formation of base pairs with the IGS extension results in formation of the P1 extension (P1ext), highlighted in red. Together the P1 and P1 extension (red) are called the extended P1 duplex. (C) The secondary structure for the Tetrahymena L-21T ribozyme (left), L-16T ribozyme (center), and the L-16T ribozyme with an oligonucleotide substrate (S) that forms the base pairs in the P1 extension (right). Definitions as given in B for the Azoarcus ribozymes.