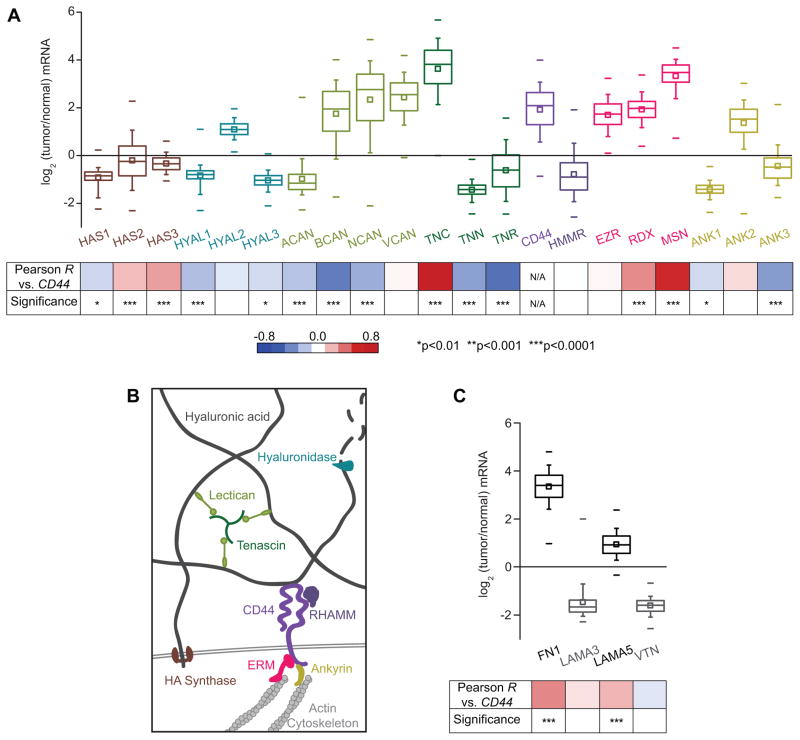
Figure 1. CD44 is frequently overexpressed in glioblastoma multiforme tumors, and correlated with misregulation of other proteins involved in CD44-mediated pathways.
(A) Transcriptomic analysis of The Cancer Genome Atlas (TCGA) microarray data: HA synthases (HAS); hyaluronidases (HYAL); the lecticans aggrecan (ACAN), brevican (BCAN), neurocan (NCAN), and versican (VCAN); the tenascins tenascin-C (TNC), tenascin-N (TNN), and tenascin-R (TNR); CD44; RHAMM (HMMR); the ERM proteins (EZR, RDX, MSN), and the ankyrins (ANK). Percentiles are represented by boxes (25, 50, 75), whiskers (10, 90), and dashes (1, 99). Bottom panel: analysis of gene expression correlation with CD44. Red-blue heat map indicates value of Pearson product-moment correlation coefficient between CD44 expression and each gene, and p values indicate the statistical significance of the correlation. (B) Expression of proteins relevant to hyaluronic acid (HA)-based brain ECM and associated downstream signaling. HA synthase proteins synthesize HA, while hyaluronidase enzymes degrade HA. Other ECM components that interact with HA include lecticans, which bind to both tenascins and HA to form a mesh-like matrix. CD44 and RHAMM are the main HA receptors. Intracellularly, CD44 links to the actin cytoskeleton through the ERM (ezrin-radixin-moesin) family proteins, and ankyrins. (C) Analysis of expression of genes encoding other extracellular matrix components previously determined to be key for glioblastoma multiforme progression: fibronectin (FN1), laminin α-5 (LAMA5), laminin α-3 (LAMA3), and vitronectin (VTN).