FIGURE 3.
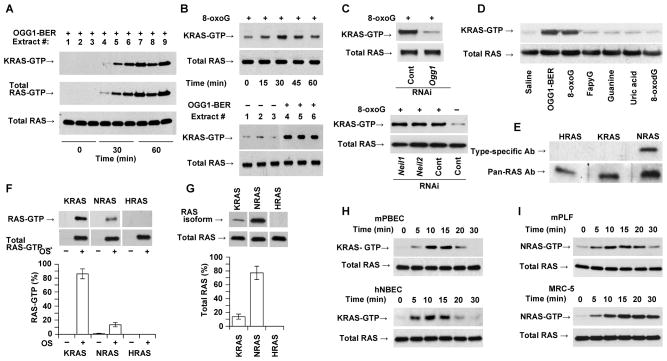
OGG1-BER and its product 8-oxoG induce activation of KRAS. (A) OGG1-BER-induced activation of KRAS in the lungs as a function of time. At indicated times, lung extracts were prepared and changes in GTP-bound RAS were determined by active RAS pull-down assays. Subtype specific Ab to K-RAS and pan-RAS Ab was used to determine KRAS-GTP and total RAS levels, respectively (Materials and Methods). (B) KRAS activation after 8-oxoG challenge (time course, upper panels) and upon initiation of OGG1-BER (lower panels, 30 min time point is shown) in individual lungs. GTP-bound and total RAS levels were determined as described in legend to 3A. (C) Activation of KRAS is OGG1- (upper panel), but not NEIL1- or NEIL2-expression dependent (lower panel) in 8-oxoG-challenged airways. GTP-bound RAS levels were determined at 30 min post-challenge. (D), OGG1-BER and 8-oxoG challenge, but not FapyG, guanine, uric acid, or 8-oxodG activates RAS (KRAS) in the lungs. The 30 min time point is shown. GTP-bound RAS was determined as described in the legend to 3A. (E) Expression of NRAS, but not KRAS or HRAS in resident alveolar macrophages. Extracts of macrophages from unchallenged lungs (n = 6) were tested by WB using subtype-specific Abs. (F) GTP-bound RAS protein levels in pooled lung extracts (n = 5) after 8-oxoG challenge, and graphical depiction of the percentages of K-, N- and HRAS-GTP in total RAS-GTP. (G) Expression of RAS subtypes in lung. K-, N- and HRAS levels (upper panels) are shown by subtype-specific Abs. Lower panel shows graphical depiction of their percentages. Total RAS level is shown by pan-RAS Ab. In F and G, band intensities were quantified using Image J (ver. 1.44) software. Expression percentages were calculated using MS Excel. (H) KRAS activation in mouse (mPBEC) and human (hNBEC) bronchial airway epithelial cells upon 8-oxoG exposure. (I) Activation of N-RAS upon 8-oxoG exposure in mouse (mPLF) and human (MRC5) lung fibroblast cells. In A–I, 250 μg extract was utilized for assessing RAS-GTP levels; 20 μg for total RAS. In B (upper panel),C,D,F, and G, lungs were challenged with 60 μL saline containing 0.0005 mg per kg 8-oxoG. In H and I, cells were challenged with 10 μM of 8-oxoG solution in medium. In A and B (lower panels) oxidative DNA damage was generated by challenging the airways with glucose oxidase (1 mU in 60 μl saline). HRAS and KRAS, mammalian homolog of Harvey and Kirsten sarcoma virus oncogen; NRAS neuroblastoma RAS viral oncogene homolog; NEIL1 and 2, Nei (endonuclease VIII)-like DNA glycosylase-1 and -2; mPBEC, mouse primary bronchial epithelial cells; mPLF, mouse primary fibroblasts; hNBEC, human normal bronchial epithelial cells; MRC5, human diploid lung fibroblasts. FapyG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; 8-oxodG, 8-oxoguanine deoxynucleoside; 8-oxoG, 7,8-dihydro-8-oxoguanine.