Figure 1.
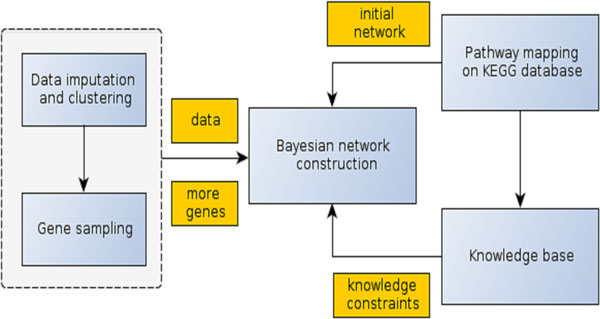
The main components of the metabolic pathway construction method and their relationships. The central component is the Bayesian network construction method. Starting from an initial metabolic pathway generated by mapping expressed genes to the KEGG pathway database, it stochastically resamples metabolic networks by drawing reactions and relations based on gene expression data and the reaction/relation knowledge base. The reaction/relation knowledge base is comprised of individual reactions and relations present in any pathway in a pathway database like KEGG regardless of which pathway or species they come from. By using all the observed reactions and relations as knowledge restraints to guide sampling, the method can more effectively and efficiently reconstruct metabolic pathways in three aspects: (1) refining an initial pathway that is largely complete; (2) reconstructing a large portion of an initial pathway when it is largely incomplete; and (3) constructing a completely de novo pathway when the initial pathway has very few or no reactions / relations.
