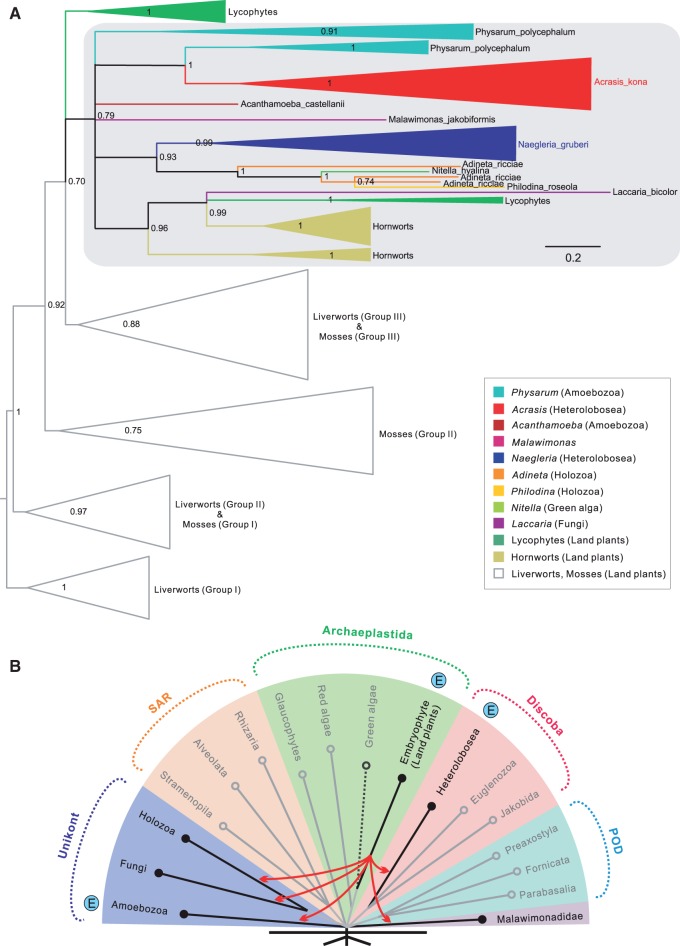
Fig. 7.—
Phylogeny of DYW-type PPR proteins in the eukaryote species. (A) Bayesian phylogeny of the E/E+/DYW domain of the DYW-type PPR proteins. Taxa include a broad sampling of early branching land plants (Liverworts, Mosses, Hornworts, and Lycophytes), and those outside land plants (Heterolobosea [Acrasis kona, Naegleria gruberi], Amoebozoa [Acanthamoeba castellanii, Physarum polycephalum], Metazoa [Adineta ricciae, Philodina roseola], Fungi [Laccaria bicolor], Charophyta [Nitella hyaline], Malawimonas jakobiformis). Values correspond to posterior probabilities; all nodes with posterior probability less than 0.7 are collapsed. Taxon labels are color-coded according to the key at the bottom right. The strong phylogenetic affinity of the only known green algal sequence (N. hyaline) for rotifers may indicate possibly transcriptomic data contamination as reported in (Laurin-Lemay et al. 2012). A detailed list of taxa is shown in supplementary figure S7, Supplementary Material online. (B) A schematic tree showing the phylogenetic distribution of DYW-type PPR proteins in the major eukaryote lineages. Lineages with experimental evidence of C-to-U type RNA editing are indicated with “E.” Arrows indicate the probable HGT directions from land plants to the other eukaryotes. Dotted line indicates the doubtful presence of DYW-type PPR proteins in N. hyaline.