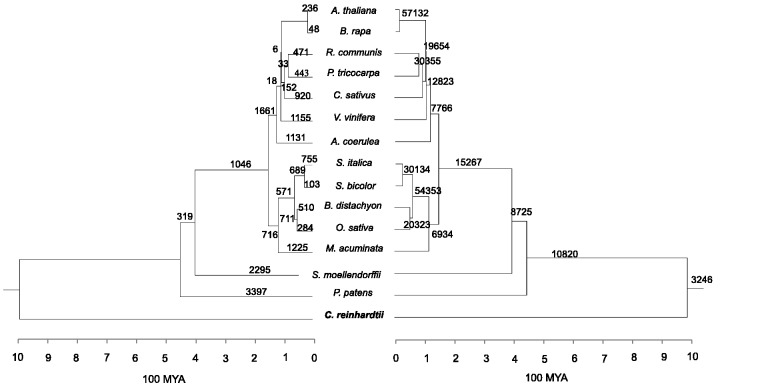
Fig. 5.—
The lineage-specific loss of ancestral CNSs and the number of CNSs found from all pairwise searches. Left panel: The lineage-specific loss of ancestral CNSs. The values on branches represent the number of CNSs lost on that specific branch. The reference genome (Ch. reinhardtii) used for this analysis is highlighted in green. Right panel: Number of CNSs found from all pairwise searches. CNSs between all pairs of species were determined to have an overall comprehensive view on gain of noncoding conservation. These pairwise analyses consider the union of all CNSs. The number on each node reflects the gain of CNSs obtained through pairwise searches. These CNSs are common to each group of species and therefore are likely to be found in outgroup species.