Abstract
Background
Small GTPases of the Rho family are critical regulators of various cellular functions including actin cytoskeleton organization, activation of kinase cascades and mitogenesis. For this reason, a major objective has been to understand the mechanisms of Rho GTPase regulation. Here, we examine the function of a novel protein, Scambio, which shares homology with the DH-PH domains of several known guanine nucleotide exchange factors for Rho family members.
Results
Scambio is located on human chromosome 14q11.1, encodes a protein of around 181 kDa, and is highly expressed in both heart and skeletal muscle. In contrast to most DH-PH-domain containing proteins, it binds the activated, GTP-bound forms of Rac and Cdc42. However, it fails to associate with V14RhoA. Immunofluorescence studies indicate that Scambio and activated Rac3 colocalize in membrane ruffles at the cell periphery. In accordance with these findings, Scambio does not activate either Rac or Cdc42 but rather, stimulates guanine nucleotide exchange on RhoA and its close relative, RhoC.
Conclusion
Scambio associates with Rac in its activated conformation and functions as a guanine nucleotide exchange factor for Rho.
Background
Rho family members including Rac, Cdc42, and Rho are essential regulators of various cellular processes such as actin cytoskeleton reorganization, mitogenesis, activation of kinase cascades, transcriptional activation, and stimulation of DNA synthesis [1-3]. Interestingly, these small GTPases have been implicated in oncogenesis, through multiple approaches [4-10]. Like all members of the Ras superfamily, Rho proteins function as molecular switches. They characteristically cycle between an active, GTP-bound state and an inactive, GDP-bound state. Switching between these states is mediated by guanine nucleotide exchange factors (GEFs) which promote the exchange of bound GDP for GTP and thereby activate the small GTPase [1]. The Dbl family of GEFs for Rho GTPases are characterized by the presence of a Dbl homology (DH) domain in tandem with a Pleckstrin homology domain (PH). While the DH domain catalyzes guanine nucleotide exchange, the PH domain is essential for membrane localization via phospholipid binding [11]. The prototype DH-PH domain-containing protein, the Dbl proto-oncogene, was originally isolated as a transforming gene from a diffuse B-cell lymphoma, and was later shown to release GDP from Cdc42 [11]. Furthermore, other Dbl-like GEFs such as Vav, Tiam, Ost, and Dbs have also been demonstrated to cause cellular transformation via their activation of Rho family proteins or through the amplification of signaling cascades [5,6,9,11]. Therefore, the study of these regulators is likely to reveal crucial details of cellular homeostasis and its disruption in cancer.
To date, three Rac proteins with an overall homology of around 90% have been identified in man. Amongst these, Rac2 is hematopoietic specific and involved in the oxidative burst, whereas Rac1 and Rac3 are ubiquitously expressed [1,12]. To elucidate the cellular functions of Rac3 in relation to its downstream effectors, we have performed a yeast two-hybrid screen using constitutively active Rac3 as bait. Here we report the identification of a novel protein that functions as both a binding partner for activated Rac and a guanine nucleotide exchange factor for Rho.
Results
Clone 78-3 represents a gene located on human chromosome 14q11.1 that is highly expressed in heart and skeletal muscle
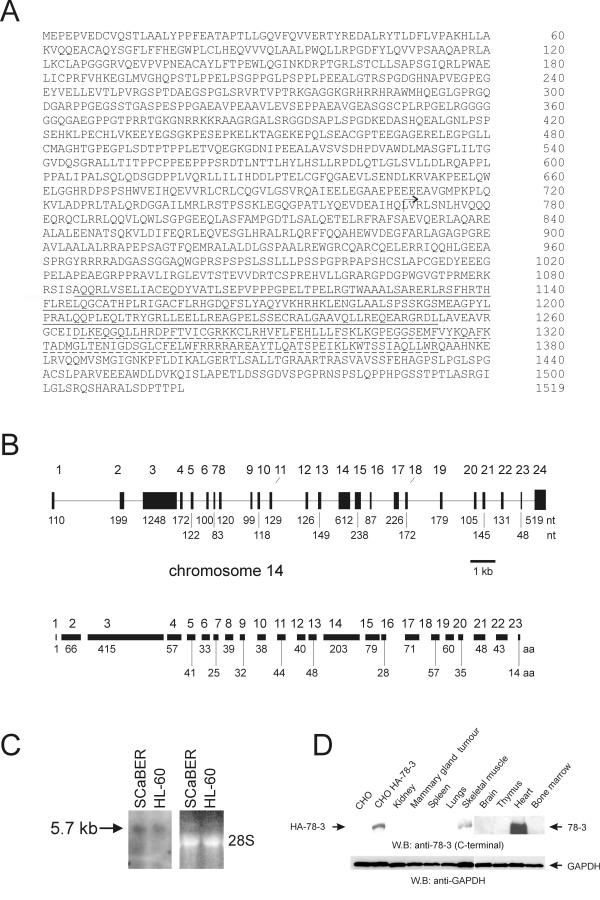
To identify downstream targets of Rac3, we performed a yeast two-hybrid screen of a human placental cDNA library using constitutively active V12Rac3, with an S189 mutation at the carboxyl-terminal end to prevent lipid modification and membrane targeting, as bait [13]. One gene was isolated twice independently as clones 78-3 and 54-5. Clone 78-3 included a stop codon at its carboxyl-terminus, whereas clone 54-5 contained an open reading frame segment of the same gene (also see Fig. 4B). We then isolated a full-length cDNA clone, from a human kidney carcinoma (A498) cell line cDNA library. The open reading frame of that clone (clone 4) predicted a protein of 1519 amino acids length, with a calculated molecular mass of 181 kDa (Fig. 1A) of which the amino acid sequence (accession number BAB84883) was previously deposited in Genbank by the Kazusa DNA Research Institute.
Figure 4.
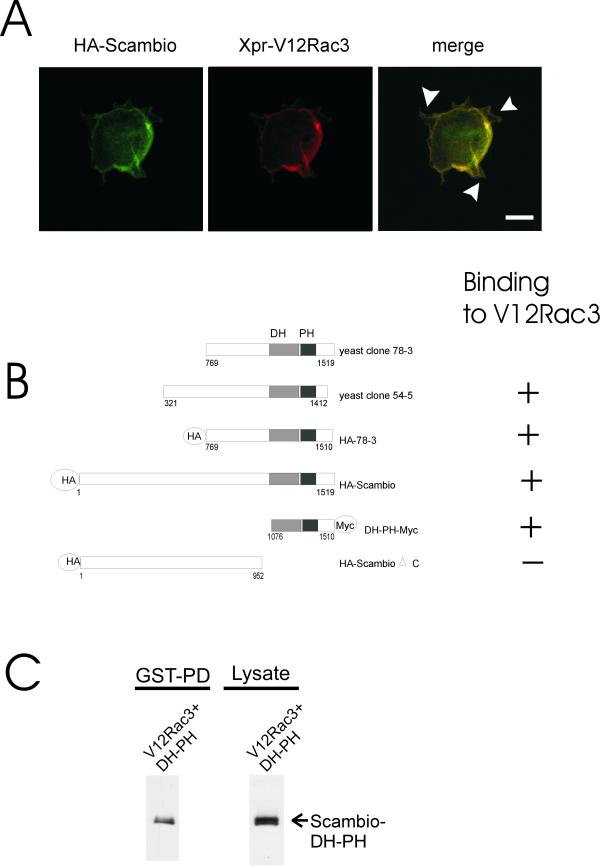
Activated Rac3 colocalizes with Scambio to membrane ruffles and binds to residues 1076–1412 of Scambio (A) NIH3T3 cells were co-transfectedwith HA-tagged Scambio and Xpress-tagged V12Rac3. Fibronectin adhered cells were stained for FITC-HA-Scambio (left panel), and Cy3-Xpress-V12Rac3 (middle panel), or merged (right panel). Arrowheads in the right panel point to the numerous ruffles where HA-Scambio and Xpress-V12Rac3 colocalize. Confocal images (× 100), Bar = 20 μm. One cell is shown that is representative of many others obtained in more than two independent experiments. (B) Schematic representation of the clones tested for binding to V12Rac3. Abbreviations used include; HA, hemagglutinin tag; DH, Dbl homology domain; and PH, pleckstrin homology domain. The DH and PH domains are shaded grey and black, respectively. (C) Activated Rac3 binds to a region encompassing the DH-PH domain of Scambio. The DH-PH domain-containing construct including a C-terminal Myc tag was co-transfected with GST-V12Rac3. Binding of this domain to GST-V12Rac3 was assessed on a GST pull-down of the lysate followed by an anti-Myc Western blot.
Figure 1.
Cloning of Scambio (A) Deduced amino acid sequence of Scambio. The DH (amino acids 1085–1253) and PH (amino acids 1265–1372) domains are underlined with solid and dashed lines, respectively. The original yeast two-hybrid positive cDNA starts at amino acid 769, as indicated with an arrow. (B) Schematic representation of the human Scambio gene structure. The size of each of the 24 exons is indicated in nucleotides below the top schematic. The number of amino acid residues encoded by each exon is shown beneath the bottom schematic. (C) Northern blot analysis of human Scambio expression. RNAs are as indicated. The filter was hybridized to a cDNA probe and washed to 2.5 × SSC at 65°C (left panel). An ethidium bromide-stained gel serves as a loading control (right panel). (D) Western blot analysis of lysates from various mouse tissues. HA-Sabi (Scambio), vector control transfected CHO-K1 lysates and tissue lysates as indicated (top panel) were immunoblotted with antibodies that recognize the C-terminal domain of 78-3. Blots were also incubated with anti-GAPDH antibodies as a loading control (bottom panel).
A panel of somatic cell hybrid DNAs that each retain one intact human chromosome was used to localize 78-3 to chromosome 14 (data not shown). The gene spans 19.2 kb and includes 24 exons (Fig. 1B). The first exon encodes the 5'-untranslated region and the first amino acid residue. Exon 24 contains only 3'-untranslated sequences.
To examine if we had isolated a complete cDNA, Northern blot analysis was performed on RNA from the human cell lines HL-60 (promyelocytic leukemia) and SCaBER (bladder carcinoma). Using the entire cDNA insert of clone 4 as probe, we detected a single mRNA of around 5.7 kb in both cell lines, in close agreement with the combined sizes of our cDNAs (Fig. 1C). Database searches with the human cDNA sequence revealed that homologues of this protein have been evolutionarily conserved in mouse. RT-PCR analysis performed on total RNA from 13 mouse tissues including heart, lung, brain, liver, kidney, spleen, bladder, intestine, ovaries, testis, stomach, small intestine and muscle, demonstrated that this gene is ubiquitously expressed (data not shown).
This analysis did not provide information with respect to relative abundance levels of the product encoded by 78-3. We therefore raised polyclonal antisera against a C-terminal domain of 78-3. To further study the relative abundance of this protein in various tissues, Western blot analysis was performed using lysates from multiple mouse tissues (Fig. 1D). CHO-K1 cells transfected with either vector or HA-78-3 were included as a negative and positive control, respectively. We examined lysates from kidney, mammary gland tumour, spleen, lungs, skeletal muscle, brain, thymus, heart, and bone marrow. As a loading control, blots were incubated with anti-GAPDH antibodies. The antibodies were able to detect a product of approximately 181 kDa in the CHO-K1 cells transfected with HA-78-3. A protein of approximately 181 kDa was also highly expressed in both heart and skeletal muscle, and was also present in the lungs, albeit at a lower level (not visible in Fig 1D).
Clone 4 encodes a DH-PH-domain containing protein, Scambio
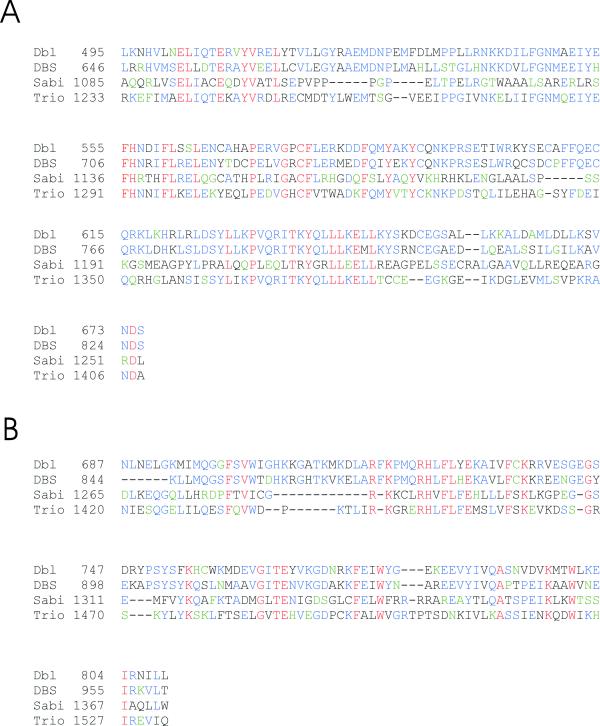
The deduced amino acid sequence of clone 4 was screened for known motifs using the ProfileScan program. This analysis showed that it contains a tandem DH-PH domain in its carboxyl-terminal end (Fig. 1A). These domains are characteristic for proteins that act as GEFs for members of the Rho family of small GTPases. Therefore, we propose to name this protein Scambio (scambio = "exchange" in Italian), abbreviated as Sabi. A BLAST database search showed that the Scambio DH and PH domains are most closely related to those of Dbl, Dbs and Trio. However, Scambio is clearly more distantly related to this group than Dbl, Trio and Dbs are to each other. Dbl and Sabi share 24% and 33% homology in their DH and PH domains, respectively (Fig. 2A and 2B).
Figure 2.
Sequence alignment of the DH and PH domains (A) DH domains. Residues 1085–1253 of Scambio (Sabi) were aligned with similar sequences of Dbl, Trio and Dbs. (B) PH domains. The PH domains of Dbl, Trio, and Dbs were aligned with residues 1265–1372 of Scambio (Sabi). Conserved amino acid residues are blue (identical residues in some aligned sequences, but not all), green (conservative substitutions) or red (identical residues in all aligned sequences). Non-homologous residues are shown in black.
Scambio binds to activated Racs or Cdc42
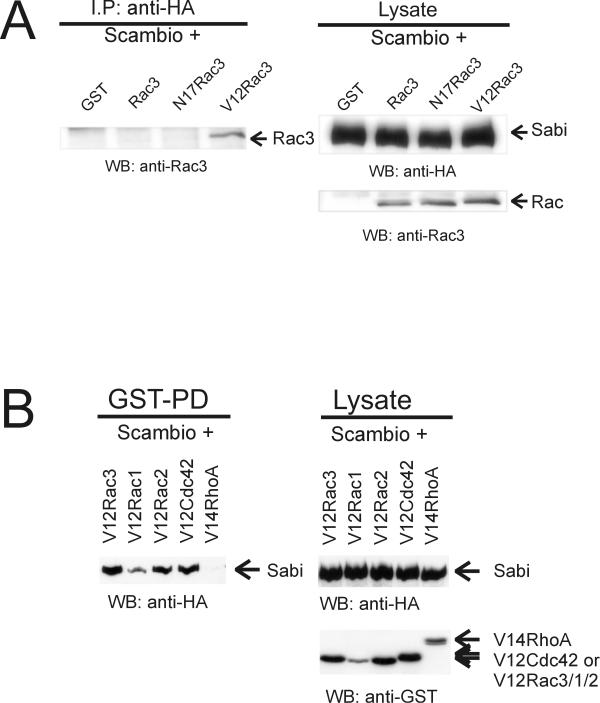
The original yeast two-hybrid positives of Scambio were isolated using constitutively active V12S189Rac3 as bait. Clone 78-3 did not interact with wild-type (S189Rac3) Rac3 in yeast (data not shown). Since the original clones encoded only part of Scambio, it was important to examine if Rac would interact with the complete protein. Therefore, we co-expressed full length, HA-tagged Scambio together with GST-tagged Rac3, N17Rac3, or V12Rac3 in CHO-K1 cells. Complex formation was evaluated via immunoprecipitation using a monoclonal anti-HA antibody and immunoblotting using polyclonal anti-Rac3 antibodies. As shown in Figure 3A (left panel), Scambio specifically complexed with the V12Rac3 mutant but not with N17Rac3, wild type Rac3 or the GST control. Thus, the interaction between Rac3 and Scambio is GTP-dependent.
Figure 3.
Scambio interacts specifically with the constitutively activated forms of Rac1, Rac2, Rac3 and Cdc42, but not with V14RhoA (A)CHO-K1 cells were transfected with HA-Sabi and GST only or GST-tagged wild type Rac3, N17Rac3, or V12Rac3. Scambio was immunoprecipitated using monoclonal anti-HA antibodies and complexed GST-tagged Rac3 mutants were detected using polyclonal Rac3 antibodies. Western blot analysis is shown of 7.5 μg of lysates (right panel) or immunoprecipitates from 0.4 mg of lysates (left panel). (B) COS-1 cells were transfected with HA-Scambio and the indicated constitutively active, GST-tagged small GTPases. GST-tagged proteins were pulled down using glutathione-agarose beads. Scambio was detected using anti-HA antibodies and GST-tagged GTPases using anti-GST antibodies. Western blot analysis is shown of 10 μg of lysate (right panel) or pull-down from 0.5 mg of lysate (GST-PD, left panel). The sizes of RhoA and Cdc42 are larger because these proteins also contain a Myc tag.
To determine whether Scambio binds to other Rho family proteins, COS-1 cells were transfected with HA-Scambio and the GST-tagged, GTP-bound forms of Rac3, Rac1, Rac2, Cdc42 and RhoA. The small GTPases were precipitated using glutathione agarose and the presence of Scambio was investigated using HA antibodies. As shown in Figure 3B, Scambio clearly interacted with all three constitutively active Rac proteins as well as with constitutively active Cdc42. However, it did not complex with GTP-bound V14RhoA. These data indicate that Scambio interacts with Rac/Cdc42 in vivo when these proteins are in their GTP-bound conformation.
Active Rac3 colocalizes with Scambio and binds within a region containing its DH-PH domain
We next investigated in which location Scambio and Rac interact within the cell. We expressed HA-tagged Scambio together with activated Xpress-tagged Rac3 in NIH3T3 cells that were plated on fibronectin. After 24 hrs, the locations of V12Rac3 and Scambio were examined using immunofluorescence and confocal microscopy. As shown in Figure 4A, middle panel, V12Rac3 localized prominently to the cell periphery and was especially concentrated in actin-based membrane ruffles. Similarly, Scambio was present at the same locations, as indicated in the individual (left panel) and merged image (Fig. 4A, right panel, arrowheads).
We also examined which domain in Scambio mediates the interaction with activated Rac3. The finding that full-length Scambio, 78-3, and 54-5 all bind to V12Rac3 already delineated the binding region to residues 769-1412 (shown in Fig 4B). A construct including residues 1–952 failed to bind V12Rac3, indicating that the N-terminal region of Scambio does not mediate Rac3 binding. Surprisingly, a small segment containing residues 1076–1510 did clearly interact with V12Rac3 (Fig. 4C). Thus, V12Rac3 binds to a region in Scambio encompassed by residues 1076–1412, which includes the DH-PH domain.
Scambio does not activate Rac and Cdc42
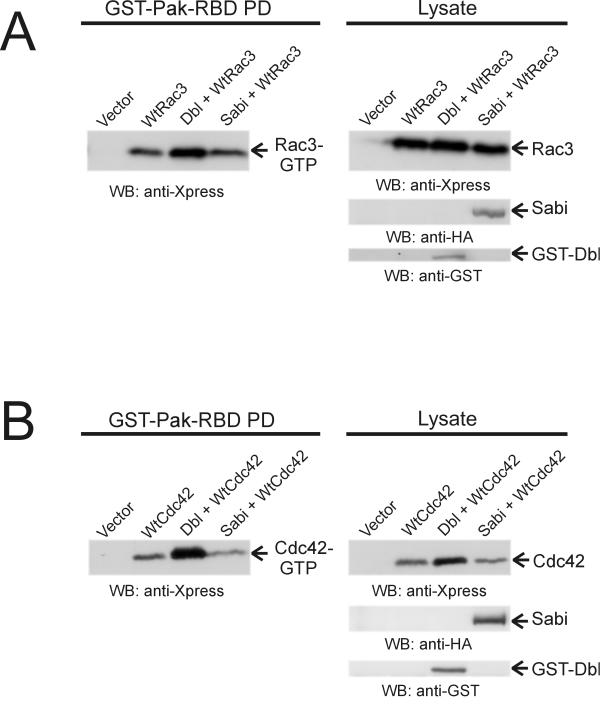
The finding that Scambio contains a DH-PH domain suggests that it functions as a guanine nucleotide exchange factor for Rho family members. To assess whether Scambio could increase the levels of activated Rac3 and Cdc42, we employed a recombinant GST-fusion protein containing the isolated GTP-dependent binding domains of the Rac and Cdc42 effector Pak1 [14]. CHO-K1 cells were transiently transfected with either Xpress-Rac3 or Xpress-Cdc42 or were co-transfected with HA-Sabi. Cells were serum-starved in 0.1% FCS for 24 hrs to induce quiescence. As a positive control, GST-tagged oncogenic Dbl (the Dbl-DH-PH domain) was co-expressed with either Xpress-Rac3 or Xpress-Cdc42. The GST-pull downs, which represent the GTP-bound fraction of the small GTPase, were immunoblotted with anti-Xpress antibodies. As shown in Fig. 5A and 5B, we were unable to detect increased levels of Rac-GTP or Cdc42-GTP upon expression with Sabi, whereas in the positive control samples, elevated levels of GTP-Rac and GTP-Cdc42 were clearly present when they were co-expressed with Dbl. This result was not unexpected. Since Rac and Cdc42 bind to Scambio in their GTP-bound conformation, Scambio is unlikely to act as an exchange factor for these small GTPases.
Figure 5.
Scambio does not stimulate guanine nucleotide exchange on Rac and Cdc42 (A) Cells were transiently transfected with either vector, wild type Rac3 (Xpress-tagged), wild type Rac3 and Dbl, or wild type Rac3 and Sabi. The GST-pull downs (left panel) were immunoblotted with anti-Xpress antibodies. Lysates (right panel) were immunoblotted with anti-Xpress, anti-HA, and anti-GST antibodies to detect Xpress-tagged Rac3, HA-Sabi, and GST-Dbl, respectively. (B) Cells were transfected with either vector alone, wild type Cdc42 (Xpress-tagged), wild type Cdc42 and Dbl or wild type Cdc42 and Sabi. The GST-pull downs for Cdc42 (left panel) and lysates (right panel) were treated as described for Rac3.
Scambio activates RhoA and RhoC
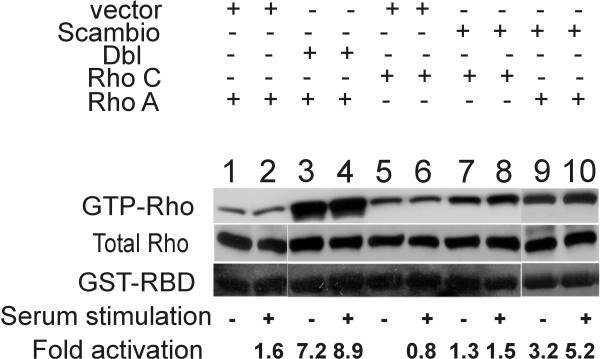
As mentioned above, V14RhoA does not bind to Scambio. This suggests that Scambio could function as a GEF for RhoA or other Rho subfamily members. To address this, we utilized a GST-fusion protein containing the Rhotekin Rho-binding domain as an affinity reagent in a pull-down assay, as described by Ren et al. [15] to determine whether Scambio could increase the level of GTP-bound RhoA in CHO-K1 cells. CHO-K1 cells were transiently transfected with either Xpress-RhoA, Xpress-RhoC or were co-transfected with HA-Scambio. Cells were serum-starved in 0.1% FCS for 24 hrs to induce quiescence, and immediately prior to lysis, cells were stimulated with 10% FCS for one minute or were further serum deprived. As a positive control, GST-tagged oncogenic Dbl was co-expressed with Xpress-RhoA. The GST-pull downs (Fig. 6, upper panel) and lysates (middle panel) which represent GTP-Rho and total Rho, respectively, were immunoblotted with anti-Xpress antibodies.
Figure 6.
Scambio stimulates guanine nucleotide exchange on RhoA and RhoC Cells were transfected with RhoA and vector (lanes 1, 2), with RhoA and Dbl (lanes 3, 4), with RhoC and vector (lanes 5, 6), with RhoC and Scambio (lanes 7, 8) or with RhoA and Scambio (lanes 9, 10). Cells were stimulated with either 10% fetal calf serum for one minute (indicated as + below the panel) or were further serum-starved immediately prior to lysis (indicated as -). The amount of GTP-Rho and total Rho was quantitated by immunoblotting with anti-Xpress antibodies on the pull-down (upper, GTP-Rho) or 1.7% of the total cellular lysate (middle, total Rho). Equal application of GST-RBD in the Rhotekin pull down assay is shown by staining of the gel with Coomassie Brilliant Blue (bottom, GST-RBD). The fold activation was determined by normalizing the GTP-Rho levels to total Rho levels and was measured relative to the level of activation seen for the control, using densitometric quantification. Similar results were obtained in more than two other independent experiments.
As expected, the expression of oncogenic Dbl increased the GTP levels of co-transfected RhoA as compared to cells transfected with the vector control (Fig. 6, compare lane 1 to lanes 3 and 4). Interestingly, Scambio also clearly stimulated guanine nucleotide exchange on RhoA: an increase in Rho-GTP levels of 3.2 and 5.2-fold was found upon serum-starvation and serum stimulation respectively, in comparison with vector and RhoA transfected cells (Fig. 6, compare lane 1 to lanes 9 and 10). We also tested the activity of Scambio towards RhoC, a small Rho family GTPase, which is closely related to RhoA but has been less intensively studied. We found that Scambio was able to stimulate the activation of RhoC relative to the control (compare lane 5 to lanes 7 and 8) when HA-Scambio and Xpress-RhoC were co-expressed. These data demonstrate that Scambio acts as a guanine nucleotide exchange factor for both RhoA and RhoC.
Scambio stimulates guanine nucleotide exchange on endogenous Rho
In the experiments described above, Scambio activated both RhoA and RhoC when these small GTPases were overexpressed. To determine whether Scambio also acts as a GEF on physiologically relevant levels of Rho proteins, we assayed for increased levels of endogenous GTP-Rho in the presence of Scambio.
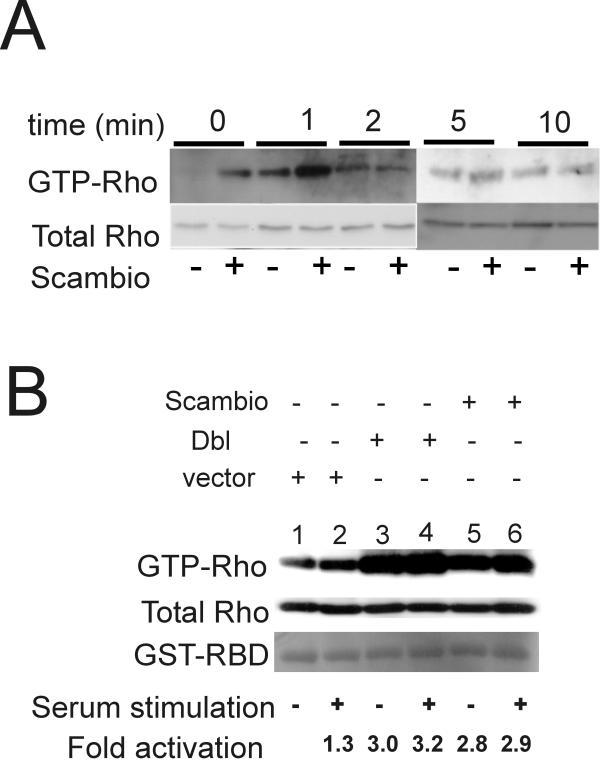
CHO-K1 cells transfected with either vector or Scambio were serum-starved, then stimulated with 10% fetal calf serum (FCS) for varying durations of time prior to assaying for levels of GTP-bound Rho. Rho activation was detected with a monoclonal antibody directed against an epitope common to different Rho proteins. As shown in Figure 7A, increases in Rho-GTP levels stimulated by Scambio after serum stimulation were transient, reaching maximal levels after one minute. We also compared the ability of Scambio to promote GTP-loading on endogenous Rho to that of oncogenic Dbl. As shown in Figure 7B, expression of Scambio caused substantial increases in the levels of endogenous Rho, albeit at a slightly lower level than that of the Dbl oncogene. These findings, in conjunction with those described above, indicate that Scambio acts as a GEF for both overexpressed and endogenous Rho.
Figure 7.
Scambio activates endogenous Rho (A) Kinetics of Rho activation by Scambio. CHO-K1 cells were transfected with either vector (indicated as a – below the panel) or with Scambio (indicated with a +), serum-starved and then stimulated with 10% serum for the number of minutes indicated above the panel. (B) Comparison of activation of endogenous Rho by Dbl and by Scambio. Cells were transfected with vector (lanes 1, 2), Dbl (lanes 3, 4) or Scambio (lanes 5, 6) and the levels of activated Rho determined as described in the legend to Fig. 6. The amount of GTP-Rho was quantitated by immunoblotting using a monoclonal antibody that recognizes an epitope common to Rho. Similar results were obtained in more than two other independent experiments.
Discussion
In this study, we have characterized Scambio, a novel nucleotide exchange factor of the Dbl family. Scambio contains a tandem DH-PH domain, the classical hallmark of a GEF for Rho family proteins. Within the Dbl family, some members act exclusively on specific small GTPases, whereas others are more promiscuous [16]. Our data indicate that Scambio functions as a guanine nucleotide exchange factor for RhoA and RhoC, but does not activate Rac or Cdc42. Thus, Scambio belongs to the class of Rho-GEFs with specificity for a subset of Rho-related small GTPases.
RT-PCR analysis indicated that Scambio is ubiquitously expressed. However, this non-quantitative assay did not allow us to determine relative expression levels. Therefore, we raised antisera and performed Western blot analysis of multiple mouse tissue lysates. This revealed that the Scambio protein is highly expressed in heart and skeletal muscle, and, to a lesser extent, in the lungs. This finding suggests that it has an important function in these tissues. Interestingly, substantial evidence indicates that RhoA is crucial to heart development and physiology [17-21]. RhoA has been shown to be highly up-regulated during early heart development, is important for normal embryogenesis [18], and has been implicated in the regulation of hypertrophic cardiac muscle cell growth [19,22]. RhoA also controls serum response factor (SRF) dependent cardiac specific gene expression via interactions with the β1-integrin signaling pathway and changes in actin cytoskeleton dynamics [17,20]. Furthermore, at least one RhoGEF specific to cardiac cells has already been identified. The p63Rho GEF, a RhoA specific, Dbl-like guanine nucleotide exchange factor, for example, has been shown to induce stress fiber formation in fibroblasts and cardiac myoblasts [21]. However, further experimentation will be necessary to determine if Scambio is also involved in cardiac development and contractility.
Scambio is unlike the majority of Dbl family members in that it can bind to activated GTP-bound Rac and Cdc42. Interestingly, the closely related GEF, Ost, which activates both RhoA and Cdc42, also binds to activated Rac1. The downstream effects of this binding are not known, but it was suggested that Ost functions as a Rac effector and links pathways involving Rac, Cdc42 and RhoA [23]. GEF-H1 is a second GEF capable of binding to the GTP-bound form of small Rho family GTPases. Unlike Scambio, it binds both activated Rac and Rho, and is additionally able to activate them. It is thought that this GEF may function to transport and activate Rac at microtubules [24,25]. The Lfc protein which binds to Rac in a nucleotide-independent manner but does not activate it, has also been suggested to function to recruit Rac to microtubuli [26].
We have not found evidence that Scambio colocalizes with microtubuli (LH, unpublished observations). However, Scambio does contain a PH domain. These domains can function as regulated membrane-binding modules that bind to inositol lipids and respond to upstream signals by targeting the host proteins to their proper cellular location [27]. Therefore, the finding that activated Rac and Scambio precisely colocalize at the cell periphery in ruffles of cells that are adhering to fibronectin suggests that Scambio recruits activated Rac to a specific subcellular location.
The activity of many Dbl family GEFs is regulated. For example, Vav is activated by tyrosine phosphorylation [28,29]. We investigated the possible activation of Scambio by overexpressing it with a constitutively active tyrosine kinase, Bcr/Abl [30], but did not detect increased GEF activity (data not shown). Hyperosmotic stress has been reported to activate Rho in a kidney tubular epithelial cell line [31]. In concordance with these results, we could detect Rho activation in CHO-K1 cells upon hypertonic treatment, but this did not increase the Scambio-mediated Rho activation. Cold stress also failed to increase Rho activation through Scambio (data not shown).
We found that Scambio had detectable GEF activity when expressed in serum-starved cells without any stimulation by external factors. Interestingly, stimulation of cells that had been serum-starved, using fetal calf serum, caused a very rapid and transient increase in the exchange factor activity of Scambio, indicating that it is regulated by a component of serum such as growth factors or lysophosphatidic acid (LPA). Since Scambio has been demonstrated to activate both RhoA and RhoC and because many cancers are characterized by deregulation of mitogenic signals via such small GTPases, it is possible that this exchange factor plays a role in certain malignancies. It will therefore be of interest to examine if any mutations can constitutively activate the GEF activity of Scambio towards Rho.
Conclusions
Scambio is encoded by a novel gene that is highly expressed in heart and skeletal muscle. It binds to Rac and Cdc42 in a GTP dependent manner, but does not activate them. Scambio functions to promote the activation of RhoA and RhoC, by acting as a guanine nucleotide exchange factor. Further studies will be important in elucidating the mechanism by which Scambio integrates these dual functions and in understanding how Scambio might link pathways that signal through Rac, Cdc42, and Rho.
Methods
Yeast two-hybrid screen
We screened a human placental cDNA library (MATCHMAKER, Clontech) with V12S189Rac3 as previously described [13]. One of 10 positives recovered in this screen was clone 78-3, which was isolated independently twice. Candidate positive cDNAs from the yeast two-hybrid screen were subcloned into pSK (Stratagene) and sequenced. The 3.1 kb cDNA clone 78-3 encodes 750 amino acids (residues 769–1519 of Scambio) and has 50-bp of 3'-untranslated sequences. cDNA clone 54-5, with a 3.2 kb insert, encodes 1077 amino acid residues (residues 321–1412 of Scambio). We screened a human kidney carcinoma (A498) cell line cDNA library using clone 78-3 as a probe to obtain a full-length cDNA. Two positives were isolated of 5.2 kb (clone 4) and 5.7 kb (clone 11). The latter had a longer 3'-untranslated region and a deletion, which caused a frame shift. Clone 4 contained the complete coding region, including a Kozak sequence, CCATGG, flanking the ATG initiator codon and is referred to as full-length Scambio. The clone also contains 100 bp of 5'-noncoding and 530 bp of 3'-untranslated sequences. The amino acid sequence of a clone identical to Scambio, isolated from a human spleen cDNA library, has been reported (locus BAB84883; gi: 18676462; nt sequence AK074057, gi: 18676461). This gene is located on a contig on chromosome 14q11.1 (NT_026437). Our original yeast clone (78-3) was digested with EcoRIxXhoI, and subcloned into pSG5. This construct contains an N-terminal HA-epitope tag from pACT-2. HA-78-3 expresses an 84-kDa protein. We subcloned the full-length HA-tagged Scambio into pSG5 and pIRESneo (Clontech). The calculated size of the protein product is 181 kDa. Additional deletion mutants include HA-Scambio C (1–952) in pIRESneo (110 kDa) and Scambio-DH-PH-Myc in pcDNA (50 kDa).
Plasmids
The plasmids pAS2-1/V12S189Rac3, pAS2-1/S189Rac3, pLEF/GST-wtRac3, pLEF/GST-V12Rac3, pLEF/GST-N17Rac3, pLEF/GST-V12Rac1, pLEF/GST-V12Rac2, pLEF/GST-V12Cdc42, pLEF/GST-V14RhoA, and pcDNA3.1/Xpress-V12Rac3 have been described previously [13,32]. Wild type RhoA and RhoC in pcDNA3.1 were from the Guthrie Research Institute. pcDNA3.1/RhoA and pcDNA3.1/RhoC were digested with EcoRIxXbaI, and subcloned into pcDNA3.1 (Invitrogen), which provides an Xpress-tag. For pIRESneo/HA-Scambio, full length HA-Scambio was subcloned into pSG5 and a 5.2 kb EcoRI fragment from this construct was inserted into pIRESneo (Clontech). Oncogenic Dbl was a gift from Dr. S.A. Aaronson (Mount Sinai School of Medicine, New York, NY). pZIP-Dbl was digested with BamHI, and a 1.8 kb fragment was inserted into pLEF [33]. GST-RBD was kindly provided by Dr. M.A. Schwartz (The Scripps Research Institute, La Jolla, CA). GST-Pak-RBD was a gift from Dr. Ulla Knaus (The Scripps Research Institute, La Jolla, CA).
Northern blotting, RT-PCR, and multiple tissue Western blotting
Total cell line RNA (15 μg) from the human bladder carcinoma SCaBER and promyelocytic leukemia HL-60 (ATCC) was run on a guanidine thiocyanate-agarose gel, blotted and hybridized using standard protocols. Blots were washed to 2.5 × SSC at 65°C. For RT-PCR of total mouse RNAs (1 μg per reaction), we selected as forward primer (in human, located in exon 2): 5'-CGCCCTTTGAAGCCACAGCC-3' and reverse primer (in human, in exon 3): 5'-TCCTGTACCCGCCCACCTCC-3', which, in human are separated by an intron. These primers correspond approximately to amino acid residues 19–133 in the human sequence and are predicted, based on the human sequence, to give a 345 bp product. Tissue lysates in SDS sample buffer obtained from mouse kidney, mammary gland tumour, spleen, lungs, skeletal muscle, brain, thymus, heart, and bone marrow were immunoblotted with antibodies that recognize the C-terminal domain of 78-3 (anti-78-3, 1:500). These polyclonal antibodies were raised in rabbit against a C-terminal region of 78-3 including amino acid residues 952–1265 expressed as a GST fusion protein in pGEX-3X. CHO-K1 cells transfected with 6 μg of either vector or HA-78-3 were included as a negative and positive control, respectively. As a loading control, blots were incubated with monoclonal anti-GAPDH antibodies (Chemicon, 1:10,000).
Interaction of small GTPases with Scambio in eukaryotic cells
CHO-K1 cells (ATCC) were transiently transfected with a total of 6 μg DNA, with full length, HA-tagged Scambio together with GST tagged wild type Rac3, N17Rac3, or V12Rac3. As a negative control, cells were transfected with GST alone. Scambio was immunoprecipitated from 0.4 mg of lysates using monoclonal anti-HA antibodies (USC core facility, 1:1000) and complexed GST-tagged Rac3 mutants were detected using polyclonal Rac3 antibodies ([12], 1:1000). Western blot analysis was performed using 7.5 μg of lysate.
COS-1 cells (ATCC) were transiently transfected with 5.5 μg of pSG5/HA-tagged-Scambio and 0.5 μg of GST-fusion protein-expressing pLEF using Lipofectamine. pLEF plasmids included pLEF/GST-V12Rac1, pLEF/GST-V12Rac2, pLEF/GST-V12Rac3, pLEF/GST-V12Cdc42, pLEF/GST-V14RhoA, or pLEF-vector plasmid. Transfected cells were lysed in Triton-lysis buffer (25 mM sodium phosphate, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μM pepstatin, and 1 mM Na3VO4) and cleared lysates (0.5 mg) were incubated with glutathione agarose beads as described [13]. GST-containing proteins were immobilized on glutathione agarose, and the presence of Scambio and GST-fusion proteins were detected with anti-HA (USC core facility, 1:1000) and polyclonal anti-GST (1:2000) antibodies. Proteins were visualized using ECL (Amersham).
Immunocytochemistry
NIH3T3 cells (ATCC) were transfected with a total of 6 μg of plasmid DNA encoding either HA-tagged full-length Scambio or with Xpress-tagged V12Rac3 using Lipofectamine (Invitrogen). Twenty-four hrs post transfection, cells were trypsinized and allowed to adhere to fibronectin (Sigma)-coated chamber slides (Nalge Nunc) for 24 hrs. Transfected cells were washed twice with phosphate buffered saline (PBS), fixed with 2% paraformaldehyde for 15 minutes, and permeabilized with 0.2% Triton X-100 in PBS for 5 minutes. Cells were then washed three times with PBS, twice with PBS/1% BSA, and blocked with 3% normal goat serum (Jackson Laboratories) in PBS for 30 minutes. Cells were immunostained with rabbit polyclonal anti-HA antibodies (1:400, Santa Cruz) or monoclonal anti-Xpress (1:400, Invitrogen) antibodies for 1 hr and washed four times with PBS. After incubation with FITC-(1:50, Zymed), or Cy3-labeled (1:200, Jackson) secondary antibodies for 30 minutes, cells were washed three times with PBS, mounted using Vectashield (Vector Laboratories), and analyzed with a Leica TCG SP confocal microscope, or a Leica DM RA fluorescence microscope.
Purification of GST-Pak-RBD and GST-RBD
The isolated GTP-dependent binding domains (amino acids 70–132) of the Rac and Cdc42 effector Pak1 (p21 activated kinase) [14] were used to affinity-precipitate activated GTP-Rac3 and GTP-Cdc42. GST-Pak-RBD purification was similar to that described by Benard et al. [14].
The Rhotekin Rho binding domain (RBD, amino acids 7–89) was used to affinity purify GTP-Rho. GST-RBD purification was essentially as described by Ren et al. [15]. Cells grown to an absorbance of 0.8 (A600) were induced for 3 hrs by the addition of 0.5 mM isopropyl B-D-thiogalatopyranoside (IPTG). Cells were suspended in 20 ml of ice-cold lysis buffer (50 mM Tris, pH 7.5, 5 mM MgCl2, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 10 μg/ml aprotinin and leupeptin, and 1 mM PMSF), sonicated and insoluble material removed by centrifugation. The GST-RBD protein was bound to glutathione agarose beads (Sigma) and the lysate-bead mixture was then washed six times in 5 ml of wash buffer (50 mM Tris, pH 7.5, 5 mM MgCl2, 150 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 10 μg/ml aprotinin and leupeptin, and 0.1 mM PMSF). After removing remaining supernatant, the beads were resuspended in 2 ml wash buffer and 10% glycerol and purified GST-RBD was stored on beads in aliquots at -80°C.
Rac3 and Cdc42 GTPase activation assays
CHO-K1 cells were seeded onto 10 cm2 tissue culture dishes in Dulbecco's modified Eagle's medium (DMEM) + 10% FCS. After 24 hrs, cells were transiently transfected with either Xpress-Rac3 or Xpress-Cdc42 or were co-transfected with HA-Scambio or vector. As a positive control, cells were co-transfected with Xpress-Rac3 or Xpress-Cdc42 plus GST-Dbl-DH-PH. After 18 hrs, cells were serum-starved in 0.1% FCS/DMEM for 24 hrs to induce quiescence. Cells were washed twice in ice-cold PBS (-/-) and harvested by scraping into 500 μl of lysis buffer (25 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM NaCl, 1% Igepal, 5% (v/v) glycerol, 1 mM Na3VO4) [14]. Cells were incubated on ice for 20 min. Following centrifugation at 13,000 g for 10 min (4°C), 450 μl of supernatant was incubated with 7.5 μl (30 μg) of GST-Pak-RBD precoupled to glutathione agarose beads (Sigma) at 4°C for one hr. An aliquot of supernatant was used to represent total protein levels. Beads were washed 4 times with 1 ml lysis buffer (without 1 mM Na3VO4). Finally, GST-Pak-RBD pull down samples or lysates were separated by SDS-PAGE and were analyzed by immunoblotting with monoclonal anti-Xpress antibodies (Invitrogen, 1:5000) to detect overexpressed Rho GTPases, monoclonal anti-HA (USC core facility, 1:1000), and polyclonal anti-GST antibodies (1:2000). Proteins were visualized using ECL (Amersham) and developed on Hyperfilm (Kodak).
RhoA and RhoC GTPase activation assays
CHO-K1 cells were seeded onto 10 cm2 tissue culture dishes in Dulbecco's modified Eagle's medium (DMEM) + 10% FCS. After 24 hrs, cells were transiently transfected with either Xpress-RhoA or Xpress-RhoC, or were co-transfected with HA-Scambio or vector. As a positive control, cells were co-transfected with Xpress-RhoA plus GST-Dbl-DH-PH. When assaying for endogenous Rho, cells were transfected with vector, HA-Scambio or GST-Dbl-DH/PH alone. After 18 hrs, cells were serum-starved in 0.1% FCS/DMEM for 24 hrs to induce quiescence and immediately prior to experimentation cells were either stimulated with 10% FCS/DMEM for one minute or were further starved. Cells were washed twice in ice-cold TBS and harvested by scraping into 600 μl of 50 mM Tris, pH 7.2, 10 mM MgCl2, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 10 μg/ml aprotinin and leupeptin, and 1 mM PMSF. 750 μg-1 mg of cell lysates were clarified by centrifugation at 13,000 g for 10 min at 4°C. The supernatant was then rotated with ~30 μg of GST-RBD precoupled to glutathione agarose beads (Sigma) at 4°C for one hr. Beads were washed four times in 600 μl of 50 mM Tris, pH 7.2, 10 mM MgCl2, 150 mM NaCl, 1% Triton X-100, 10 μg/ml aprotinin and leupeptin, and 0.1 mM PMSF. Finally, GST-RBD pull downs and lysates were eluted in 2 × SDS sample buffer, separated by SDS-PAGE, and analyzed by immunoblotting with monoclonal anti-Xpress antibodies (Invitrogen, 1:5000) to detect overexpressed Rho GTPases or with a monoclonal anti-Rho antibody (BD Transduction Laboratories, 1:250) that recognizes an epitope common to different Rho proteins to detect endogenous Rho. Proteins were visualized using ECL (Amersham) and developed on Hyperfilm (Kodak). Densitometric analysis of Rho activation levels was performed using UnScan It Software (Silk Scientific).
Abbreviations
ATCC, American Type Culture Collection; bp, base pairs; DTT, dithiothreitol; ECL, enhanced chemiluminescence; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GST, glutathione-S-transferase; HA, hemaggluttinin; hr, hour; nt, nucleotide; PBS, phosphate buffered saline; PMSF, phenylmethylsulfonyl fluoride; RT-PCR, reverse transcription polymerase chain reaction; SSC, saline sodium citrate; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TBS, tris buffered saline
Authors' contributions
CC wrote the manuscript and performed the Rho activation experiments (Fig. 6 and Fig. 7). BH performed the Western blot analysis of Scambio expression levels, immunoprecipitations (Fig. 1D, Fig. 2, Fig. 3A, Fig. 5), and contributed to the experimental design and technical implementation of the experiments. LH performed the initial yeast two-hybrid screen, Northern blot, analyzed binding to Rac and Cdc42, and performed the Scambio deletion mutant and immunohistochemistry analyses (Fig. 1B,1C, Fig. 3B, Fig. 4). DS cloned the full-length Scambio cDNAs, sequenced them, and performed the RT-PCR analysis and made subclones (Fig. 1A). JG contributed to overall experimental design. NH was responsible for the design and implementation of all the experiments and the final manuscript.
Acknowledgments
Acknowledgements
We thank Dr. A. Hall for generously providing Myc-tagged V12Cdc42 and V14RhoA plasmids, Dr. S.A. Aaronson for Dbl, Dr. M.A. Schwartz for GST-RBD, Dr. U.G. Knaus for GST-Pak-RBD, and Dr. F. Rudert for the pLEF-vector. We thank Dr. G. McNamara of the CHLARI Image core for his assistance with the confocal microscopy analysis. *This work was supported by PHS grants CA90321 and CA50248 (NH) and by the T.J. Martell Foundation (NH and JG).
Contributor Information
Christina Curtis, Email: christina@rustycup.com.
Bianca Hemmeryckx, Email: hemmeryc@usc.edu.
Leena Haataja, Email: LHaataja@mednet.ucla.edu.
Dinithi Senadheera, Email: Dsenadheera@chla.usc.edu.
John Groffen, Email: jgroffen@chla.usc.edu.
Nora Heisterkamp, Email: heisterk@hsc.usc.edu.
References
- Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Malek RL, Behbahani B, Castellone MD, Lee NH, Gutkind JS. Identification of H-Ras, RhoA, Rac1 and Cdc42 responsive genes. Oncogene. 2003;22:2689–2697. doi: 10.1038/sj.onc.1206364. [DOI] [PubMed] [Google Scholar]
- Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Cerione RA, Manor D. Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J Biol Chem. 1999;274:23633–23641. doi: 10.1074/jbc.274.33.23633. [DOI] [PubMed] [Google Scholar]
- Zohn IM, Campbell SL, Khosravi-Far R, Rossman KL, Der CJ. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/S0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- Reuther GW, Lambert QT, Booden MA, Wennerberg K, Becknell B, Marcucci G, Sondek J, Caligiuri MA, Der CJ. Leukemia-associated Rho guanine nucleotide exchange factor, a Dbl family protein found mutated in leukemia, causes transformation by activation of RhoA. J Biol Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- Knaus UG. Rho GTPase signaling in inflammation and transformation. Immunol Res. 2000;21:103–109. doi: 10.1385/IR:21:2-3:103. [DOI] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/S0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a novel member of the Rho family. J Biol Chem. 1997;272:20384–20388. doi: 10.1074/jbc.272.33.20384. [DOI] [PubMed] [Google Scholar]
- Haataja L, Groffen J, Heisterkamp N. Identification of a novel Rac3-interacting protein C1D. Int J Mol Med. 1998;1:665–670. doi: 10.3892/ijmm.1.4.665. [DOI] [PubMed] [Google Scholar]
- Benard V, Bohl BP, Bokoch GM. Characterization of Rac and Cdc42 Activation in Chemoattractant-stimulated Human Neutrophils Using a Novel Assay for Active GTPases. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- Ren XD, Schwartz MA. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/S0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Arai A, Spencer JA, Olson EN. STARS, a striated muscle activator of Rho Signaling and serum response factor-dependent transcription. J Biol Chem. 2002;277:24453–24459. doi: 10.1074/jbc.M202216200. [DOI] [PubMed] [Google Scholar]
- Kaarbo M, Crane DI, Murrell WG. RhoA is highly up-regulated in the process of early heart development of the chick and important for normal embryogenesis. Dev Dyn. 2003;227:35–47. doi: 10.1002/dvdy.10283. [DOI] [PubMed] [Google Scholar]
- Sah VP, Minamisawa S, Tam SP, Wu TH, Dorn GW, Ross J, Jr, Chien KR, Brown JH. Cardiac-specific overexpression of RhoA results in sinus and atrioventricular nodal dysfunction and contractile failure. J Clin Invest. 1999;103:1627–1634. doi: 10.1172/JCI6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Wang L, Carson JA, Agan JE, Imanaka-Yoshida K, Schwartz RJ. Beta-1 integrin and organized actin filaments facilitate cardiomyocyte-specific RhoA-dependent activation of the skeletal alpha-actin promoter. FASEB J. 2001;15:785–796. doi: 10.1096/fj.00-026com. [DOI] [PubMed] [Google Scholar]
- Souchet M, Portales-Casamar E, Mazurais D, Schmidt S, Leger I, Javre JL, Robert P, Berrebi-Bertrand I, Bril A, Gout B, Debant A, Calmels TP. Human p63 RhoGEF, a novel RhoA-specific guanine nucleotide exchange factor, is localized in cardiac sarcomere. J Cell Sci. 2002;115:629–640. doi: 10.1242/jcs.115.3.629. [DOI] [PubMed] [Google Scholar]
- Aoki H, Izumo S, Sadoshima J. Angiotensin II activates RhoA in cardiac myocytes: a critical role of RhoA in angiotensin II-induced premyofibril formation. Circ Res. 1998;82:666–676. doi: 10.1161/01.res.82.6.666. [DOI] [PubMed] [Google Scholar]
- Horii Y, Beeler JF, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- Krendel K, Zenke FT, Bokoch GM. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat Cell Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- Glaven JA, Whitehead I, Bagrodia S, Kay R, Cerione RA. The Dbl-related protein, Lfc, localises to microtubules and mediates the activation of Rac signaling pathways in cells. J Biol Chem. 1999;274:2279–2285. doi: 10.1074/jbc.274.4.2279. [DOI] [PubMed] [Google Scholar]
- Vanni C, Mancini P, Gao Y, Ottaviano C, Guo F, Salani B, Torrisi MR, Zheng Y, Eva A. Regulation of proto-Dbl by intracellular membrane targeting and protein stability. J Biol Chem. 2002;277:19745–19753. doi: 10.1074/jbc.M111025200. [DOI] [PubMed] [Google Scholar]
- Han J, Das B, Wei W, Van Aelst L, Mosteller RD, Khosravi-Far , Westwick JK, Der CJ, Broek D. Lck regulates Vav activation of members of the Rho family of GTPases. Mol Cell Biol. 1997;17:1346–1353. doi: 10.1128/mcb.17.3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structure basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/S0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Groffen J. Philadelphia-positive leukemia: a personal perspective. Oncogene. 2002;21:8536–8540. doi: 10.1038/sj.onc.1206080. [DOI] [PubMed] [Google Scholar]
- Ciano-Oliveira CD, Sirokmany G, Szaszi K, Arthur WT, Masszi A, Peterson M, Rotstein OD, Kapus A. Hyperosmotic stress activates Rho: differential involvement in Rho kinase-dependent MLC phosphorylation and NKCC activation. Am J Physiol Cell Physiol. 2003;285:C555–C566. doi: 10.1152/ajpcell.00086.2003. [DOI] [PubMed] [Google Scholar]
- Senadheera D, Haataja L, Groffen J, Heisterkamp N. The small GTPase Rac interacts with ubiquitination complex proteins Cullin-1 and CDC23. Int J Mol Med. 2001;8:127–133. doi: 10.3892/ijmm.8.2.127. [DOI] [PubMed] [Google Scholar]
- Rudert F, Visser E, Gradl G, Grandison P, Shemshedini L, Wang Y, Grierson A, Watson J. pLEF, a novel vector for expression of glutathione S-transferase fusion proteins in mammalian cells. Gene. 1996;169:281–282. doi: 10.1016/0378-1119(95)00820-9. [DOI] [PubMed] [Google Scholar]