Abstract
BACKGROUND/AIMS
Biological and epidemiological data suggest that vitamin D levels may influence cancer development. Several single nucleotide polymorphisms have been described in the vitamin D receptor (VDR) gene in association with cancer risk. We aimed to investigate the association of VDR gene polymorphisms with hepatocellular carcinoma (HCC) development in chronic hepatitis C patients.
METHODS
In a cross-sectional, hospital-based setting, 340 patients (201 chronic hepatitis, 47 cirrhosis and 92 HCC) and 100 healthy controls receiving VDR genotyping (bat-haplotype: BsmI rs1544410 C, ApaI rs7975232 C and TaqI rs731236 A) were enrolled.
RESULTS
Patients with HCC had a higher frequency of ApaI CC genotype (P = 0.027) and bAt[CCA]-haplotype (P = 0.037) as compared to control subjects. There were no differences in BsmI and TaqI polymorphisms between two groups. In patients with chronic hepatitis C, HCC subjects had a higher frequency of ApaI CC genotype and bAt[CCA]-haplotype than those with chronic hepatitis (P = 0.001 and 0.002, respectively) and cirrhosis (P = 0.019 and 0.026, respectively). After adjusting age and sex, logistic regression analysis showed that ApaI CC genotype (odds ratio: 3.02, 95% confident interval: 1.65-5.51) was independently associated with HCC development.
CONCLUSION
VDR ApaI polymorphism plays a role in the development of HCC among chronic hepatitis C patients. Further explorations of this finding and its implications are required.
Introduction
Hepatitis C virus (HCV) infection is one of the major public health problems worldwide [1]. Chronic HCV infection is characterized by a high rate of progression to fibrosis, chronic hepatitis, leading to cirrhosis and ultimately to hepatocellular carcinoma (HCC) [2], [3], [4]. Although the relationship between HCV and the development of HCC is well established, the pathogenetic mechanism of hepatocarcinogenesis, including host- and viral-related factors, is still unknown. It is prudent to affirm that differences in the incidence rates and the strong gender distribution in HCC are not entirely due to differences in the exposure to the causative agents [5], [6]. Of great importance, genetic factors can also contribute, particularly gene polymorphisms of inflammatory cytokines and growth factor ligands and receptors [7].
Vitamin D is involved in the metabolism of skeleton as a systemic hormone but also has important roles in the regulation of host immune responses, fibrogenesis and development of cancer through vitamin D receptor (VDR) [8], [9], [10], [11], [12], [13], [14]. Previous data have suggested that vitamin D levels may influence cancer development. In particular, several single nucleotide polymorphisms have been described in the VDR gene, and some polymorphisms are associated with tumor occurrence [12], [13], [14], [15], [16]. For instance, VDR polymorphisms have been related to cancers of the breast, prostate, skin, colon-rectum, bladder and kidney, although with conflicting observations [12], [13], [14], [15], [16]. VDR polymorphisms have also been investigated in the context of some chronic liver diseases, such as chronic hepatitis B, primary biliary cirrhosis and autoimmune hepatitis [17], [18], [19]. In a recent published study, VDR polymorphism may be used as a molecular marker to predict the risk and to evaluate the disease severity of HCC in patients with chronic hepatitis B [20].
So far, there are limited data in the literature on the association between VDR polymorphisms and the occurrence of HCC. In this present study, we investigated the role of VDR gene polymorphisms in the susceptibility and clinicopathological status of HCC in Chinese subjects with chronic HCV infection.
Patients and Methods
Patients
From August 2011 to July 2013, a total of 340 patients with chronic HCV infection receiving long-term follow up in a single center were enrolled. They included 201 chronic hepatitis, 47 cirrhosis and 92 HCC patients. All patients were seropositive for HCV antibody (by third-generation enzyme-linked immunosorbent array (ELISA) and HCV RNA (Amplicor™, Roche Diagnostics, Branchburg, NJ, USA). Patients were excluded if they were positive for serum hepatitis B surface antigen or anti-human immunodeficiency virus antibody, or exhibited other causes of hepatocellular injury (e.g. any history of alcoholism, autoimmune hepatitis, primary biliary cirrhosis and severe nonalcoholic liver disease with metabolic syndrome). During the same period, 100 healthy volunteers were collected as controls.
Pathologic diagnoses of chronic hepatitis or cirrhosis were made by percutaneous liver biopsies according to the modified Knodell histologic activity index [21], which were analyzed by pathologists who were blind to the patients’ characteristics. Diagnosis of HCC was based on either the histopathologic findings in tumor tissues or typical HCC features of dynamic images if the nodules were larger than 1 cm in cirrhotic livers [22]. This study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethical committees of Chang Gung Memorial Hospital. All patients gave written informed consent before enrollment.
Detection of VDR Polymorphisms
The DNA was extracted from peripheral blood leukocytes using the Qiagen DNA isolation kit (Qiagen, Germany). The VDR genotype was determined by polymerase chain reaction (PCR) amplication and restriction length fragment polymorphisms (RFLP) as previously described [23]. For the detection of BsmI polymorphisms, a forward primer in exon 7 (5’-CAACCAAGACTCAAGTACCGCGTCAGTGA-3’) and a reverse primer in intron 8 (5’-AACCAGCGGAAGAGGTCAAGGG) was used. For the detection of ApaI and TaqI polymorphisms, a forward primer in exon 8 (5’-CAGAGCATGGACAGGGAGCAA) and a reverse primer in exon 9 (5’-GCAACTCCTCATGGCTGAGGTCTC) was used. The PCR products for BsmI polymorphisms were 820 base pair (bp), and for ApaI/TaqI polymorphisms they were 745 bp. The PCR mix contained 5 μL of each primer (10 pmol), 5 μL buffer, 1.5 μL MgCl2 (50 mM), 5 μL template DNA (50–100 ng), 5 μL dNTPs (2 mmol/L), Taq polymerase (MBI) 2 μL, H2O 26.5 μL. The DNA template was denatured at 95°C for 2 min. A total of 40 cycles of PCR were performed, consisting of a denaturation step for 45 sec at 94°C, an annealing step for 45 sec at optimum temperature (67°C for Apa I/Taq I and 60°C for Bsm I), and an extension reaction for 1 min at 72°C. A final extension step at 72°C for 2 min was added after the last PCR cycle.
After amplication, the PCR products were digested with BsmI, ApaI and TaqI endonucleases. Following restriction endonuclease digestion, genotyping was determined by ethidum bromide-UVB illumination of the fragments separated on gels of 2% agarose. The presence of BsmI, ApaI or TaqI restriction site was defined as the lower-case ‘b’, ‘a’ and ‘t’, respectively, and the absence of the site was defined as the upper-case ‘B’, ‘A’ or ‘T’. The BsmI restriction site resulted in two fragments (645 bp and 177 bp). Digestion with ApaI produced two fragments of 531 bp and 214 bp when the restriction site was present. Digestion with TaqI resulted in three fragments of approximately 205, 290 bp and 245 bp in the presence of TaqI polymorphic site, and in fragments of 245 and 495 bp in the absence of a TaqI polymorphic site.
Statistical Analysis
Continuous data are expressed as mean ± standard deviation, and the categorical data are expressed as number (percentage). Comparisons of differences in the categorical date between groups were performed using the chi-square test. Distributions of continuous variables were analyzed by the Student’s t-test or one-way ANOVA test with least significant difference (LSD) post-hoc correction between groups where appropriate. Stepwise logistic regression analysis was performed to assess the influence of each factor on the risk of developing HCC. All analyses were carried out using SPSS software version 15.0 (SPSS Inc., Chicago, IL). All tests were 2-tailed, and a p-value of less than 0.05 was considered statistically significant.
Results
Baseline Features of the Studied Population
The basic demographical and clinical features of the patients are shown in Table 1. The mean age of HCC patients was significantly higher than those with cirrhosis, chronic hepatitis and controls (P < 0.001). Patients with HCC had a higher male-to-female ratio than other groups (P = 0.001). There was no significant difference in BMI among these groups. The HCC subjects had lower platelet count compared to those with chronic hepatitis; whereas the platelet count was comparable between cirrhosis and HCC groups.
Table 1.
Baseline Characteristics of the Studied Population
| Con (n = 100) | CH (n = 201) | LC (n = 47) | HCC (n = 92) | P-value | |
|---|---|---|---|---|---|
| Age (years) | 52.7 ± 15.6c | 52.4 ± 11.5e | 55.9 ± 10.0f | 64.5 ± 11.6cef | <0.001 |
| Male gender (%) | 43 (43%)c | 101 (50%)e | 21 (45%)f | 64 (70%)cef | 0.001 |
| BMI (kg/m2) | − | 24.6 ± 3.6 | 25.3 ± 3.6 | 24.2 ± 3.6 | 0.239 |
| AST (U/L) | 42 ± 33abc | 77 ± 58ade | 107 ± 41bd | 102 ± 80ce | <0.001 |
| ALT (U/L) | 50 ± 53abc | 138 ± 131ae | 147 ± 76bf | 94 ± 76cef | <0.001 |
| Platelet (104/μL) | − | 18.7 ± 5.4ab | 13.7 ± 6.1a | 13.2 ± 7.1b | <0.001 |
Data are expressed as mean ± standard deviation or number (percentage).
P-value by one-way ANOVA test or x2 test; a Significant differences between Con and CH; b Significant differences between Con and LC; c Significant differences between Con and HCC; d Significant differences between CH and LC; e Significant differences CH and HCC; f Significant differences between LC and HCC with LSD post-hoc correction or x2 test.
Abbreviation: Con: healthy control; CH: chronic hepatitis; LC: liver cirrhosis; HCC: hepatocellular carcinoma; BMI: body mass index; AST: aspartate aminotransferase; ALT: alanine aminotransferase.
The Distribution of Frequencies of VDR Genotype and Haplotype
Firstly, HCC patients were compared with a control cohort of 100 healthy volunteers with regard to allelic frequency. The distribution of the alleles of BsmI, ApaI and TaqI was in accordance with the Hardy-Weinberg equilibrium in both individuals of HCC and controls (P > 0.05 for any). Patients with HCC had a higher frequency of ApaI CC genotype (P = 0.027) and bAt[CCA]-haplotype consisting of BsmI C, ApaI C and TaqI A alleles (P = 0.037) as compared to control subjects. For the BsmI and TaqI polymorphisms, no significant associations were found.
Figure 1 shows the association of VDR gene polymorphisms with the disease severity in chronic HCV infection. Patients carrying the corresponding ApaI CC genotype had a higher prevalence (34%) of HCC than those with CA (19.2%) or AA type (12.5%) (P = 0.024). In contrast, BsmI and TaqI polymorphisms were not significantly associated with disease severity of chronic HCV infection. As shown in Table 2, patients with HCC carried a higher ratio of ApaI CC genotype compared to those with chronic hepatitis (P = 0.001) or cirrhosis (P = 0.026).
Figure 1.
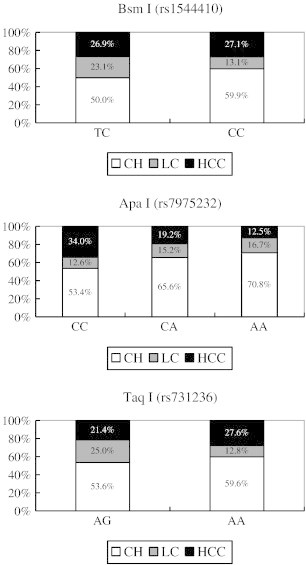
Association of VDR gene polymorphisms with the disease severity in chronic HCV infection. Bsm I (P = 0.343), Apa I (P = 0.024) and Taq I (P = 0.195).
Table 2.
The Distribution of Frequencies of VDR Genotype and Haplotype Among Controls and Different Clinical Stages of Chronic HCV Infection
| Con (n = 100) | CH (n = 201) | LC (n = 47) | HCC (n = 92) | |
|---|---|---|---|---|
| BsmI (rs1544410) | ||||
| TT | 0 (0%) | 0 (0%) | 0 (0%) | 0 (%) |
| TC | 11 (11%) | 13 (6%) | 6 (13%) | 7 (8%) |
| CC | 89 (89%) | 188 (94%) | 41 (87%) | 85 (92%) |
| T vs. C allele | 11:189 | 13:389 | 6:88 | 7:177 |
| ApaI (rs7975232) | ||||
| CC | 55 (55%)a | 102 (51%)b | 24 (51%)c | 65 (71%)abc |
| CA | 40 (40%)d | 82 (41%)e | 19 (40%) | 24 (26%)de |
| AA | 5 (5%) | 17 (8%) | 4 (9%) | 3 (3%) |
| C vs. A allele | 150:50f | 286:116g | 67:27h | 154:30fgh |
| TaqI (rs731236) | ||||
| GG | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| AG | 14 (14%) | 15 (7%) | 7 (15%) | 6 (7%) |
| AA | 86 (86%) | 186 (93%) | 40 (85%) | 86 (93%) |
| G vs. A allele | 14:186 | 15:387 | 7:87 | 6:178 |
| BsmI–ApaI–TaqI | ||||
| TAG (BaT) | 10 (10%) | 12 (6%) | 5 (11%) | 5 (5%) |
| CCA (bAt) | 54 (54%)i | 102 (51%)j | 24 (51%)k | 64 (70%)ijk |
| CAA (bat) | 31 (31%) | 83 (41%)l | 15 (32%) | 21 (23%)l |
| CAG (baT) | 4 (4%) | 3 (1%) | 2 (4%) | 0 (0%) |
| TAA (Bat) | 0 (0%) | 1 (1%) | 1 (2%) | 1 (1%) |
| TCG (BAT) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) |
| TCA (BAt) | 1 (1%) | 0 (0%) | 0 (0%) | 0 (0%) |
| CCA vs. TAG & CAA | 54:41m | 102:95n | 24:20 | 64:26mn |
Data are expressed as number (percentage). P-value by x2 test or Fisher’s exact test;
Abbreviation: VDR, vitamin D receptor; Con: healthy control; CH: chronic hepatitis; LC: liver cirrhosis; HCC: hepatocellular carcinoma.
P = 0.027; bP = 0.001; cP = 0.026; dP = 0.047; eP = 0.018; fP = 0.044; gP = 0.001; hP = 0.019; iP = 0.037; jP = 0.003; kP = 0.041; lP = 0.002; mP = 0.048; nP = 0.002
Factors Associated with Developing HCC by Logistic Regression Analysis
As shown in Table 3, univariate analysis revealed that age, male gender, lower platelet count (<15 × 104/μL), the carriage of bAt[CCA]-haplotype and ApaI CC genotype were factors significantly associated with developing HCC. Stepwise logistic regression analysis showed that age (odds ratio (OR): 1.10, 95% confidence interval (CI): 1.07-1.14, P < 0.001), male gender (OR: 3.90, 95% CI: 2.07-7.35, P < 0.001), low platelet count (<15 × 104/μL)(OR: 4.20, 95% CI: 2.26-7.83, P < 0.001) and the carriage of ApaI CC genotype (OR: 2.77, 95% CI: 1.47-5.21, P = 0.002) were the independent predictors.
Table 3.
Univariate Analyses and Stepwise Multivariate Analyses of Factors Associated with the Risk of HCC in Patients with Chronic HCV Infection
| Univariate Analyses |
Stepwise Multivariate Analyses |
|||
|---|---|---|---|---|
| Odds Ratio (95% CI) | P-value | Odds Ratio (95% CI) | P-value | |
| Age (per 1 year increase) | 1.10 (1.07-1.13) | <0.001 | 1.10 (1.07-1.14) | <0.001 |
| Male gender | 2.36 (1.42-3.93) | 0.001 | 3.90 (2.07-7.35) | <0.001 |
| BMI | 0.96 (0.89-1.03) | 0.212 | ||
| Platelet (<15 × 104/μL) | 4.36 (2.59-7.34) | <0.001 | 4.20 (2.26-7.83) | <0.001 |
| CCA vs. TAG&CAA | 2.25 (1.33-3.78) | 0.002 | − | − |
| BsmI CC type | 1.01 (0.41-2.48) | 0.987 | − | − |
| ApaI CC type | 2.33 (1.40-3.89) | 0.001 | 2.77 (1.47-5.21) | 0.002 |
| TaqI AA type | 1.40 (0.55-3.56) | 0.486 | − | − |
Abbreviations: HCC: hepatocellular carcinoma; HCV, hepatitis C virus; CI, confidence interval; BMI: body mass index.
Comparison Between Chronic HCV Patients with Apa I CC Type and CA/AA Type
Since ApaI CC genotype was a significant factor associated with developing HCC, we thus compared the chronic hepatitis C patients with ApaI CC type and CA/AA type. As shown in Table 4, patients carrying ApaI CC genotype had a higher prevalence of HCC and pre-existing cirrhosis and a higher ratio of BsmI CC type and TaqI AA type as compared to those with ApaI CA/AA type.
Table 4.
Comparison Between Chronic HCV Patients with ApaI CC Type and CA/AA Type
| ApaI CC (N = 191) | ApaI CA/AA type (N = 149) | P-value | |
|---|---|---|---|
| Age (years) | 55.8 ± 12.7 | 56.6 ± 12.2 | 0.600 |
| Male gender (%) | 106 (55%) | 80 (54%) | 0.412 |
| BMI (kg/m2) | 24.6 ± 3.2 | 24.6 ± 4.1 | 0.908 |
| LC* | 85 (45%) | 50 (34%) | 0.026 |
| HCC | 65 (34%) | 27 (18%) | 0.001 |
| AST (U/L) | 83 ± 57 | 96 ± 78 | 0.139 |
| ALT (U/L) | 131 ± 133 | 123 ± 83 | 0.522 |
| Platelet (104/μL) | 16.1 ± 6.3 | 17.0 ± 6.8 | 0.153 |
| BsmI CC type | 190 (99%) | 124 (83%) | <0.001 |
| TaqI AA type | 190 (99%) | 122 (82%) | <0.001 |
Data are expressed as mean ± standard deviation or number (percentage). P-value by Student’s t-test or x2 test. *Analysis included 47 LC patients without HCC and 88 HCC patients with underlying LC.
Abbreviations: HCV: hepatitis C virus; BMI: body mass index; LC: liver cirrhosis; HCC: hepatocellular carcinoma; AST, aspartate transaminase; ALT, alanine transaminase.
Discussion
Hepatocarcinogenesis is a complex and multi-factorial process, in which both environmental and genetic features interfere and contribute to malignant transformation [24]. The identification of genetic factors related to HCC susceptibility may improve our understanding of the various biological pathways involved in hepatocarcinogenesis and as well improve the scientific basis for preventative intervention. Numerous candidate-gene studies have reported associations between single nucleotide polymorphism and the development of HCC [24], [25], [26], [27], [28]. In this study, we investigated the possible association between the VDR gene polymorphisms and HCC in a Chinese population with chronic HCV infection. Our data showed that patients with HCC had a higher frequency of ApaI CC genotype and bAt[CCA]-haplotype as compared to control subjects. Furthermore, stepwise logistic regression analyses revealed that ApaI CC genotype was an independent factor, suggesting that the ApaI C polymorphisms may be used as a molecular marker to predict the risk of HCC in the patients infected with HCV.
Association studies of several polymorphisms in the VDR gene have been performed to investigate their implication with severity of chronic liver disease [17], [18], [19], [20]. One of the common genetic variations of VDR gene is the bat-haplotype consisting of BsmI, ApaI and TaqI [29]. These genetic variations have been described as important modulators of several chronic liver diseases such as primary biliary cirrhosis and autoimmune hepatitis [18], [19]. Recently, bAt [CCA]-haplotype VDR polymorphisms have been reported to influence antiviral response to peginterferon plus ribavirin therapy in chronic hepatitis C [30], [31]. In particular, Baur et al. have demonstrated that bAt[CCA]-haplotype and ApaI CC genotype are both significantly associated with a rapid fibrosis progression rate and with the presence of cirrhosis in patients with chronic hepatitis C [32]. This result was similar to that in our study showing that patients carrying ApaI CC genotype had a higher prevalence of cirrhosis compared to those with ApaI CA/AA type if the HCC patients with pre-existing cirrhosis were enrolled for analysis. Moreover, our data further showed that ApaI C polymorphisms might not only affect the fibrosis progression and the presence of cirrhosis, but also have a direct association with HCC development in chronic HCV infection.
Two recent studies have reported the relationship between VDR gene polymorphisms and HCC development in patients with chronic HCV infection [33], [34]. Falleti et al. have demonstrated that VDR genetic polymorphisms are significantly associated with the occurrence of HCC in cirrhotic patients who underwent liver transplantation [33]. However, this relationship is more specific for patients with an alcoholic etiology, but not in those with cirrhosis of viral origin. This discrepancy could be explained by the limited case numbers in the subgroup analysis of virus-cirrhotic subjects. The other study has reported that genetic variations in CYP2R1, GC, and DHCR7 are associated with progression to HCC in patients with chronic hepatitis C according to four heterogeneous independent cohorts [34].
In this study, we cannot prove the causal relationships between genetic variations and distinct clinical phenotypes. However, the significant association between the polymorphisms in VDR which serves as the physiological target to mediate vitamin D effects and HCV-induced HCC suggests that an impaired vitamin D metabolism contributes to hepatocarcinogenesis in chronic HCV infection. Although serum vitamin D levels and history regarding vitamin D intake (dietary or supplemental) were not available, this could be justified since VDR gene variants modulate biological effects of vitamin D without influencing vitamin D plasma levels [29], [35]. In addition, 25(OH)D3 serum levels strongly fluctuate during seasons, with age, and as a consequence of numerous other conditions [8], [36].
In conclusion, the present study suggests a significant association of VDR ApaI polymorphism with the development of HCC in chronic HCV infection. The characterization of VDR genetic polymorphisms in HCV carriers may help to identify those who are at high risk of developing HCC. This observation needs to be validated in further studies.
Acknowledgement
This study was supported in part by contract grants CLRPG8B0052 and CMRPG8A0751 from Chang Gung Memorial Hospital, Taiwan.
References
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 3.Massard J, Ratziu V, Thabut D, Moussalli J, Lebray P, Benhamou Y, Poynard T. Natural history and predictors of disease severity in chronic hepatitis C. J Hepatol. 2006;44(1 Suppl.):S19–S24. doi: 10.1016/j.jhep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Hung CH, Lee CM, Wang JH, Hu TH, Chen CH, Lin CY, Lu SN. Impact of diabetes mellitus on incidence of hepatocellular carcinoma in chronic hepatitis C patients treated with interferon-based antiviral therapy. Int J Cancer. 2011;128:2344–2352. doi: 10.1002/ijc.25585. [DOI] [PubMed] [Google Scholar]
- 5.Yu MW, Chen CJ. Hepatitis B and C viruses in the development of hepatocellular carcinoma. Crit Rev Oncol Hematol. 1994;17:71–91. doi: 10.1016/1040-8428(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 6.Hung CH, Lee CM, Lu SN, Wang JH, Hu TH, Tung HD, Chen CH, Chen WJ, Changchien CS. Long-term effect of interferon alpha-2b plus ribavirin therapy on incidence of hepatocellular carcinoma in patients with hepatitis C virus-related cirrhosis. J Viral Hepat. 2006;13:409–414. doi: 10.1111/j.1365-2893.2005.00707.x. [DOI] [PubMed] [Google Scholar]
- 7.Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37:493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- 8.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 9.Peterlik M, Cross HS. Vitamin D and calcium deficits predispose for multiple chronic diseases. Eur J Clin Invest. 2005;35:290–304. doi: 10.1111/j.1365-2362.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 10.Heubi JE, Hollis BW, Specker B, Tsang RC. Bone disease in chronic childhood cholestasis. I: vitamin D absorption and metabolism. Hepatology. 1989;9:258–264. doi: 10.1002/hep.1840090216. [DOI] [PubMed] [Google Scholar]
- 11.Chiu KC, Chu A, Go V, Saad MF. Hypovitaminosis D is associated with insulin resistance and β-cell dysfunction. Am J Clin Nutr. 2004;79:820–825. doi: 10.1093/ajcn/79.5.820. [DOI] [PubMed] [Google Scholar]
- 12.Köstner K, Denzer N, Müller CS, Klein R, Tilgen W, Reichrath J. The relevance of vitamin D receptor (VDR) gene polymorphisms for cancer: a review of the literature. Anticancer Res. 2009;29:3511–3536. [PubMed] [Google Scholar]
- 13.Tang C, Chen N, Wu M, Yuan H, Du Y. Fok1 polymorphism of vitamin D receptor gene contributes to breast cancer susceptibility: a meta-analysis. Breast Cancer Res Treat. 2009;117:391–399. doi: 10.1007/s10549-008-0262-4. [DOI] [PubMed] [Google Scholar]
- 14.Raimondi S, Johansson H, Maisonneuve P, Gandini S. Review and meta-analysis on vitamin D receptor polymorphisms and cancer risk. Carcinogenesis. 2009;30:1170–1180. doi: 10.1093/carcin/bgp103. [DOI] [PubMed] [Google Scholar]
- 15.Bai YH, Lu H, Hong D, Lin CC, Yu Z, Chen BC. Vitamin D receptor gene polymorphisms and colorectal cancer risk: a systematic meta-analysis. World J Gastroenterol. 2012;18:1672–1679. doi: 10.3748/wjg.v18.i14.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onen IH, Ekmekci A, Eroglu M, Konac E, Yesil S, Biri H. Association of genetic polymorphisms in vitamin D receptor gene and susceptibility to sporadic prostate cancer. Exp Biol Med (Maywood) 2008;233:1608–1614. doi: 10.3181/0803-RM-110. [DOI] [PubMed] [Google Scholar]
- 17.Huang YW, Liao YT, Chen W, Chen CL, Hu JT, Liu CJ, Lai MY, Chen PJ, Chen DS, Yang SS. Vitamin D receptor gene polymorphisms and distinct clinical phenotypes of hepatitis B carriers in Taiwan. Genes Immun. 2010;11:87–93. doi: 10.1038/gene.2009.65. [DOI] [PubMed] [Google Scholar]
- 18.Vogel A, Strassburg CP, Manns MP. Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology. 2002;35:126–131. doi: 10.1053/jhep.2002.30084. [DOI] [PubMed] [Google Scholar]
- 19.Fan L, Tu X, Zhu Y, Zhou L, Pfeiffer T, Feltens R, Stoecker W, Zhong R. Genetic association of vitamin D receptor polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. J Gastroenterol Hepatol. 2005;20:249–255. doi: 10.1111/j.1440-1746.2005.03532.x. [DOI] [PubMed] [Google Scholar]
- 20.Yao X, Zeng H, Zhang G, Zhou W, Yan Q, Dai L, Wang X. The association between the VDR gene polymorphisms and susceptibility to hepatocellular carcinoma and the clinicopathological features in subjects infected with HBV. Biomed Res Int. 2013:953974. doi: 10.1155/2013/953974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513–1520. [PubMed] [Google Scholar]
- 22.Bruix J and Sherman M; American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Springer JE, Cole DE, Rubin LA, Cauch-Dudek K, Harewood L, Evrovski J, Peltekova VD, Heathcote EJ. Vitamin D-receptor genotypes as independent genetic predictors of decreased bone mineral density in primary biliary cirrhosis. Gastroenterology. 2000;118:145–151. doi: 10.1016/s0016-5085(00)70423-9. [DOI] [PubMed] [Google Scholar]
- 24.Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol. 2012;57:663–674. doi: 10.1016/j.jhep.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Gao SJ, Du BX, Wang JJ. Association of IL-6 polymorphisms with hepatocellular carcinoma risk: evidences from a meta-analysis. Tumour Biol. 2014;35:3551-3361. doi: 10.1007/s13277-013-1469-5. [DOI] [PubMed] [Google Scholar]
- 26.Liu K, Zhang L, Lin X, Chen L, Shi H, Magaye R, Zou B, Zhao J. Association of GST genetic polymorphisms with the susceptibility to hepatocellular carcinoma (HCC) in Chinese population evaluated by an updated systematic meta-analysis. PLoS One. 2013;8:e57043. doi: 10.1371/journal.pone.0057043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Yin Z, Cao S, Gao W, Liu L, Yin Y, Liu P, Shu Y. Systematic review and meta-analysis on the association between IL-1B polymorphisms and cancer risk. PLoS One. 2013;8:e63654. doi: 10.1371/journal.pone.0063654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Y, Liu F, Li B, Chen X, Ma Y, Yan L, Wen T, Xu M, Wang W, Yang J. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a huge systematic review and meta-analysis. Dig Dis Sci. 2011;8:2227–2236. doi: 10.1007/s10620-011-1617-y. [DOI] [PubMed] [Google Scholar]
- 29.Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Baur K, Mertens JC, Schmitt J, Iwata R, Stieger B, Frei P, Seifert B, Bischoff Ferrari HA, von Eckardstein A, Müllhaupt B. The vitamin D receptor gene bAt (CCA) haplotype impairs the response to pegylated-interferon/ribavirin-based therapy in chronic hepatitis C patients. Antivir Ther. 2012;17:541–547. doi: 10.3851/IMP2018. [DOI] [PubMed] [Google Scholar]
- 31.García-Martín E, Agúndez JA, Maestro ML, Suárez A, Vidaurreta M, Martínez C, Fernández-Pérez C, Ortega L, Ladero JM. Influence of vitamin D-related gene polymorphisms (CYP27B and VDR) on the response to interferon/ribavirin therapy in chronic hepatitis C. PLoS One. 2013;8:e74764. doi: 10.1371/journal.pone.0074764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baur K, Mertens JC, Schmitt J, Iwata R, Stieger B, Eloranta JJ, Frei P, Stickel F, Dill MT, Seifert B. Combined effect of 25-OH vitamin D plasma levels and genetic vitamin D receptor (NR 1I1) variants on fibrosis progression rate in HCV patients. Liver Int. 2012;32:635–643. doi: 10.1111/j.1478-3231.2011.02674.x. [DOI] [PubMed] [Google Scholar]
- 33.Falleti E, Bitetto D, Fabris C, Cussigh A, Fontanini E, Fornasiere E, Fumolo E, Bignulin S, Cmet S, Minisini R. Vitamin D receptor gene polymorphisms and hepatocellular carcinoma in alcoholic cirrhosis. World J Gastroenterol. 2010;16:3016–3024. doi: 10.3748/wjg.v16.i24.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange CM, Miki D, Ochi H, Nischalke HD, Bojunga J, Bibert S, Morikawa K, Gouttenoire J, Cerny A, Dufour JF. Genetic analyses reveal a role for vitamin D insufficiency in HCV-associated hepatocellular carcinoma development. PLoS One. 2013;8:e64053. doi: 10.1371/journal.pone.0064053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen CJ. Clinical practice. Vitamin D insufficiency. N Engl J Med. 2011;364:248–254. doi: 10.1056/NEJMcp1009570. [DOI] [PubMed] [Google Scholar]


