Abstract
Anorexia nervosa (AN), bulimia nervosa (BN) and obsessive-compulsive disorder (OCD) are complex psychiatric disorders with shared obsessive features, thought to arise from the interaction of multiple genes of small effect with environmental factors. Potential candidate genes for AN, BN and OCD have been identified through clinical association and neuroimaging studies; however, recent genome-wide association studies of eating disorders (ED) so far have failed to report significant findings. In addition, few, if any, studies have interrogated postmortem brain tissue for evidence of expression quantitative trait loci (eQTLs) associated with candidate genes, which has particular promise as an approach to elucidating molecular mechanisms of association. We therefore selected single-nucleotide polymorphisms (SNPs) based on candidate gene studies for AN, BN and OCD from the literature, and examined the association of these SNPs with gene expression across the lifespan in prefrontal cortex of a nonpsychiatric control cohort (N=268). Several risk-predisposing SNPs were significantly associated with gene expression among control subjects. We then measured gene expression in the prefrontal cortex of cases previously diagnosed with obsessive psychiatric disorders, for example, ED (N=15) and OCD/obsessive-compulsive personality disorder or tics (OCD/OCPD/Tic; N=16), and nonpsychiatric controls (N=102) and identified 6 and 286 genes that were differentially expressed between ED compared with controls and OCD cases compared with controls, respectively (false discovery rate (FDR) <5%). However, none of the clinical risk SNPs were among the eQTLs and none were significantly associated with gene expression within the broad obsessive cohort, suggesting larger sample sizes or other brain regions may be required to identify candidate molecular mechanisms of clinical association in postmortem brain data sets.
Introduction
Eating disorders (ED)—particularly anorexia nervosa (AN) and bulimia nervosa (BN)—are complex genetic disorders likely involving multiple genes of small effects, interacting with multiple social and environmental factors. Throughout developed nations these disorders affect roughly 0.1% of individuals across the lifespan, over 90% of whom are female.1 Psychiatric comorbidity is high in ED, and AN has the highest mortality rate of any psychiatric illness.2,3 While twin and family studies of ED have supported a strong genetic influence for these disorders,2,4 and association studies have reported a multitude of promising candidates for AN and BN, (among them BDNF, various serotonin-related genes (most commonly 5-HT2A), COMT, dopamine, leptin and cannabinoids3, 4, 5, 6, 7, 8), replication of these findings has been inconsistent,7,8 which is not entirely surprising given similar replication problems in candidates for other major psychiatric disorders. For example, a recent large-scale candidate gene association study failed to find a single significant association with illness,9 or with alternative clinical phenotypes.10 In addition, two recent genome-wide association studies (GWAS) of common SNPs and rare CNVs in over 1000 patients with AN and over 3500 pediatric controls11 and over 5500 AN cases and 21 000 controls12 also failed to detect a single SNP with genome-wide significance, suggesting that either much larger samples or other alternate approaches to GWAS may need to be considered to advance the study of psychiatric genetics in ED.11
A number of core pathological features of ED, namely obsessionality, perfectionism, anxiety, thought preoccupations, hoarding, a strong need to control one's environment and rigidity are also common to obsessive-compulsive disorder (OCD),13 and as such, a shared genetic liability and/or brain circuitry have been suggested among the obsessive psychiatric syndromes of AN, BN and OCD, or at least between AN and OCD.14 Comorbidity of AN and OCD is common, as is to a lesser extent, AN and OCPD (obsessive-compulsive personality disorder).15
Over the past 10 years, our laboratory as well as others has successfully utilized postmortem human brain tissue from healthy control subjects in a ‘genetic neuropathology' approach to study the effect of allelic variation on gene expression.16, 17, 18, 19 Genetic association at the level of the normal tissue transcriptome can provide insight into gene function, and is not confounded by clinical epiphenomena typically seen in patient samples. We have previously suggested that postmortem brain mRNA could serve as the ‘ultimate intermediate phenotype' available to examine these disorders,17,20 particularly when these data are considered in conjunction with data from in vivo studies. To our knowledge, no study has explored the possible effects of allelic variation on mRNA expression specifically for AN, BN or OCD, in either a control or psychiatric postmortem human brain sample. Associating genetic risk variants with changes in gene expression identifies candidate mechanisms by which clinical variants increase (or decrease) risk for these disorders, and can direct future in vivo and in vitro functional studies.
Given the phenotypic and genotypic overlap between ED and OCD, we set out to evaluate risk-associated genes and SNPs that have been previously reported for AN, BN and OCD. We also identified genes that were differentially expressed comparing ED and OCD patients with controls in the first postmortem human brain sample of its kind. Finally, we examined whether the risk variants could explain the differential expression through cis or trans genetic mechanisms. Our results may guide future postmortem and in vivo brain research on psychiatric cases with AN, BN and/or OCD.
Materials and methods
Study participants
Brain specimens were donated through the Offices of the Chief Medical Examiners of the District of Columbia and of the Commonwealth of Virginia, Northern District to the NIMH Brain Tissue Collection at the National Institutes of Health in Bethesda, MD, according to NIH Institutional Review Board guidelines (Protocol #90-M-0142). Audiotaped informed consent was obtained from legal next-of-kin on every case. All NIMH cases in the obsessive cohort met DSM-IV criteria for one or more lifetime Axis I diagnosis of an ED (AN, BN or ED, not otherwise specified), and/or OCD, OCPD and/or a tic disorder. Clinical data included family informant interviews with next-of-kin, retrospective psychiatric record reviews and medical examiner data including cause/manner of death, all of which were summarized in a psychiatric narrative format and reviewed by two board-certified psychiatrists. Details of the donation process and clinical diagnostic procedures are described elsewhere.21,22 Additional specimens, including 37 second-trimester fetal brain tissue samples, were obtained through the National Institute of Child Health and Human Development Brain and Tissue Bank, and six additional psychiatric specimens were obtained from the New York Brain Bank at Columbia University, University of California at Irvine Brain Bank, and University of Texas Southwest Brain Bank. Detailed demographic information on study participants are provided in Tables 1, 2 and Supplementary Table 1. Postmortem psychiatric diagnoses for the six additional psychiatric cases were ascertained according to DSM-IV, via comparable clinical procedures with a consensus psychiatric review, at each of these collaborating brain tissue collections. All postnatal nonpsychiatric control cases (N=231) were free from psychiatric diagnoses and substance abuse according to DSM-IV. Every control case had toxicology screening to exclude for acute drug and alcohol intoxication/use at the time of death, and all the fetal tissues were also screened for possible in utero drug exposure.
Table 1. Nonpsychiatric control sample demographics.
| N | Sex (% M) | Race (%) | Age (mean±s.d.) | pH (mean±s.d.) | PMI (mean±s.d.) | RIN (mean±s.d.) | |
|---|---|---|---|---|---|---|---|
| Postnatal cases | 231 | 68.4 | Cauc=47.2 AA=48.5 Other=4.3 | 32.4±20.5 | 6.5±0.3 | 30.2±15.3 | 8.2±0.8 |
| Fetal cases | 37 | 48.6 | Cauc=8.1 AA=91.9 | 17.7±1.7 weeks | — | 2.6±2.3 | 9.7±0.2 |
| All cases | 268 | 65.4 | Cauc=41.8 AA=54.5 Other=3.7 | 27.9±22.2 | 6.5±0.3 | 26.4±17.1 | 8.4±0.9 |
Abbreviations: AA, African American; Cauc, Caucasian; Other, Hispanic, Asian, Mixed Race; pH, brain pH; PMI, postmortem interval in hours; RIN, RNA integrity number.
Table 2. Obsessive psychiatric syndromes sample demographics.
| N | Sex (% F) | Age | pH | PMI | RIN | Subtype | MoodDx | Substance Dx | Suicide | Antidepr Tox | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ED | 15 | 86.7 | 40.1 ±8.1 | 6.2 ±0.2 | 32.3 ±19.5 | 7.57 ±0.88 | 7 BN 2 AN-BN 3 AN 3 NOS | 86.7% | 53.3% | 73.3% | 66.7% |
| OC/Tic | 16 | 12.5 | 47.1 ±17.3 | 6.4 ±0.3 | 26.8 ±18.1 | 7.76 ±0.90 | 9 OCD 3 Tic/Tour 3 OCPD | 75.0% | 50.0% | 50.0% | 25.0% |
| Both | 31 | 48.4 | 43.7 ±13.9 | 6.3 ±0.3 | 29.5 ±18.7 | 7.67 ±0.88 | 15 ED 16 OCD/Tic | 80.6% | 51.6% | 61.3% | 45.2% |
| Control | 102 | 20.5 | 42.5 ±17.2 | 6.6±0.3 | 27.2±14.8 | 8.3±0.69 | |||||
| Control (subset) | 30 | 23.9 | 45.8±15.6 | 6.5±0.3 | 28.7±15.3 | 8.1±0.75 |
Abbreviations:
AN, anorexia nervosa (restricting); AN-BN, anorexia nervosa, binge–purge type; AntidepressTox, percentage of cases on antidepressants at the time of death as measured by blood/brain toxicology; BN, bulimia nervosa; Cauc, Caucasian; ED, eating disorder; MoodDx, presence of comorbid Axis I mood disorder diagnosis; NOS, eating disorder not otherwise specified; OCD, obsessive-compulsive disorder; OCPD, obsessive-compulsive personality disorder; OC/Tic, obsessive-compulsive and/or tic disorder subset; pH, brain pH; PMI, postmortem interval in hours; RIN, RNA integrity number; Substance Dx, presence of comorbid Axis I substance use disorder; Tic/Tour, tic disorder or Tourette's syndrome.
All numerical cells are mean±s.d.
We measured gene expression levels in postmortem human brain dorsolateral prefrontal cortex (DLPFC) tissue in two samples—a control-only cohort (N=268) whose gene expression measurements were generated on the Illumina Human 49 K Oligo array (two-color) and have previously been published17 and a case–control cohort of 133 subjects (15 ED patients, 16 OCD patients and 102 controls) with gene expression measurements generated on Illumina HumanHT-12 v3 microarrays (one-color), and normalized with background correction, variance stabilizing transform, followed by quantile normalization.23 There were 70 subjects in the lifespan control cohort17 that were also included as controls in the case–control cohort, but had independently generated gene expression measurements from a different microarray platform.
Tissue processing
All specimens were flash-frozen and screened for macro- and microscopic neuropathological abnormalities, as previously described.21 All specimens with evidence of neurological disorders, infarcts or other cerebrovascular abnormalities were excluded from the study. Brain pH was measured, and postmortem interval (in hours) was calculated for every sample. Postmortem tissue homogenates of the prefrontal cortex (DLPFC, BA46/9) were obtained from all subjects. For all samples regardless of microarray platform, total RNA was extracted, reverse transcribed with oligo dT, T7 amplified and labeled with the Cy3 fluorescent dye. The RNA extraction process is described in detail elsewhere.17,21
Genotyping
DNA for genotyping was obtained from the cerebella of samples and performed with either the Illumina Human Hap 650v3 or 1 M Duo V3 BeadArrays. Genotypes were called using BeadExpress software. SNPs were removed if the call rate was <98% (mean call rate for this study >99%), if not in Hardy–Weinberg Equilibrium (P<0.001) in Caucasian or African American samples, or not polymorphic (MAF<0.01). We then performed genome-wide imputation using the 1000 Genomes reference panel, ShapeIt for pre-phasing of haplotypes24 and Impute2 software package,25 which ensured we had genotype data (imputed or observed) on SNPs from the literature.
SNP selection
We conducted a comprehensive literature search using PubMed, to identify all published risk-associated single-nucleotide polymorphisms (SNPs) for AN, BN and OCD with reported nominal statistical significance (P⩽0.05), and identified 105 unique candidate SNPs (some SNPs appeared twice in the literature) annotated to 44 genes. Some SNPs may be in linkage disequilibrium, but we did not enforce independent statistical signal since we queried the literature, and the results (see below) are not confounded by this. We also included 13 SNPs (Table 2 in Boraska et al.12) with the greatest evidence for association with AN12 and 9 SNPs with the greatest evidence for association with OCD26 from the largest and most recent GWAS for each disorder, for a total of 127 unique SNPs. However, we restricted our subsequent analyses to only SNPs that were observed or imputed in our data sets with a minor allele frequency (MAF) >5%. Two SNPs were not in the 1000 Genomes Reference Panel, and thus not imputed. In the control lifespan sample, there were 114 SNPs available for analysis (76 observed, 38 imputed and 11 filtered for MAF<5%), and we extracted these same 114 SNPs out of the genome-wide imputed genotype data in the case–control series (Supplementary Table 2).
Statistical analysis
We used linear regression to analyze the expression data with respect to identifying expression quantitative trait loci and genes differentially expressed with diagnosis (see Supplementary Methods). For both expression data sets (control cohort and case–control cohort), we used surrogate variable analysis to control for technical confounders like batch effects.27 In the case–control cohort, we subsampled 30 of the 103 controls to remove the confounding effect of slight tissue quality differences between the ED, OCD and control groups. Although tissue quality, measured indirectly through RNA integrity number (RIN), was generally quite high for these postmortem tissue samples (all samples with RIN>5, 96% samples with RINs>6 and 91% samples with RINs>7), its levels were nevertheless strongly associated with gene expression measurements, and slightly different between diagnostic groups (Table 2). Although removing the highest quality controls reduced our sample size, it balanced the distributions of RIN across diagnostic groups, removing the confounding effect of RIN (Supplementary Methods). Gene set enrichment analyses were performed with the Wilcoxon signed-rank test using predefined gene sets available from Kortenhorst et al.28
Results
Few obsessive psychiatric syndromes clinical risk SNPs associate with gene expression in nonpsychiatric controls
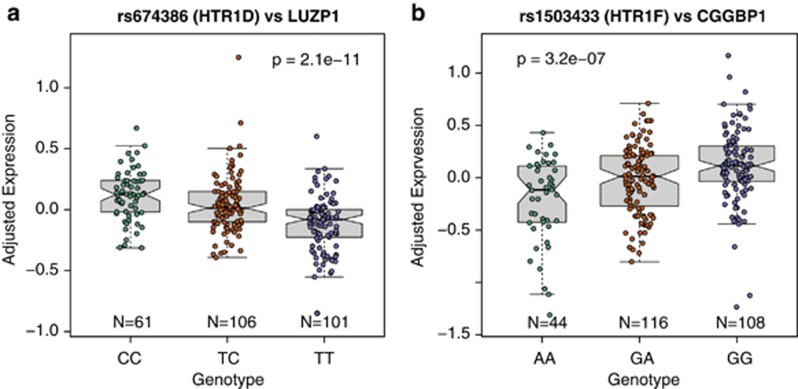
We first interrogated the potential functional relevance of the candidate ED risk SNPs by exploring their effect on gene expression in nonpsychiatric controls in the human prefrontal cortex. We performed an expression quantitative trait loci (eQTLs) analysis in postmortem brain tissue from 268 nonpsychiatric controls across the lifespan using 105 identified clinical risk SNPs for ED/OCD (see Materials and Methods) to understand how genetic variation at these loci are associated with nearby gene expression in the ‘normal' human brain. We first performed a local cis analysis—each SNP was associated with expression levels for all genes within one megabase upstream or downstream, and identified only two significant loci (Supplementary Table 3). The top cis signal involved four correlated SNPs annotated to the locus of HTR1D (rs7532266, rs674386, rs588387 and rs856510) at chr11:23521835-23551623, which was at least marginally significant in three independent studies.11,29,30 While this genetic locus contained HTR1D, the clinical risk variants were actually associated with the expression of LUZP1 (P=2.08 × 10−11, FDR=6.73 × 10−8), an adjacent gene 26 kb upstream that encodes a protein containing a leucine zipper motif (Figure 1a). Similarly, the other significant cis signal involved genetic variation in the HTR1F gene associating with the expression levels of the adjacent CGGBP1 gene (Figure 1b), a CGG triplet binding protein involved in fragile X syndrome and intellectual disability.31
Figure 1.
Top expression quantitative trait loci (eQTLs) of ED clinical risk variants associated with gene expression in nonpsychiatric controls (N=268). (a) The effect of SNP rs674386 on LUZP1 expression and (b) the effect of rs1503433 on CGGBP1 expression. Y axis is log2 expression relative to a reference pool, adjusted for surrogate variables, sex and race.
We next expanded potential genetic association to transcriptome-wide expression (for example, trans eQTLs). At this more stringent significance threshold, only the cis effect involving LUZP1 remained significant at FDR<5% in addition, there were no significant trans associations. These results therefore suggest that LUZP1 and CGGBP1 may be responsible for the clinical risk association for ED at these significant loci, rather than previously reported serotonin receptor genes HTR1D and HTR1F, respectively. We note the low rate of eQTLs among reported clinical risk SNPs—only five reported SNPs across two loci were significantly associated with local gene expression levels in the DLPFC.
Obsessive psychiatric syndromes associate with differential gene expression in the brain
We then sought to identify genes that were differentially expressed within ED patients and then within the broad obsessive cohort to identify genes associated with illness in postmortem DLPFC brain tissue. This cohort consisted of 31 cases with OCD/OCPD/Tics (N=16) and ED (N=15), compared with RIN-matched nonpsychiatric Caucasian adult controls (N=30; see Materials and Methods). First we identified gene expression differences within each illness, and then across a broader obsessive phenotype, compared with the control subjects.
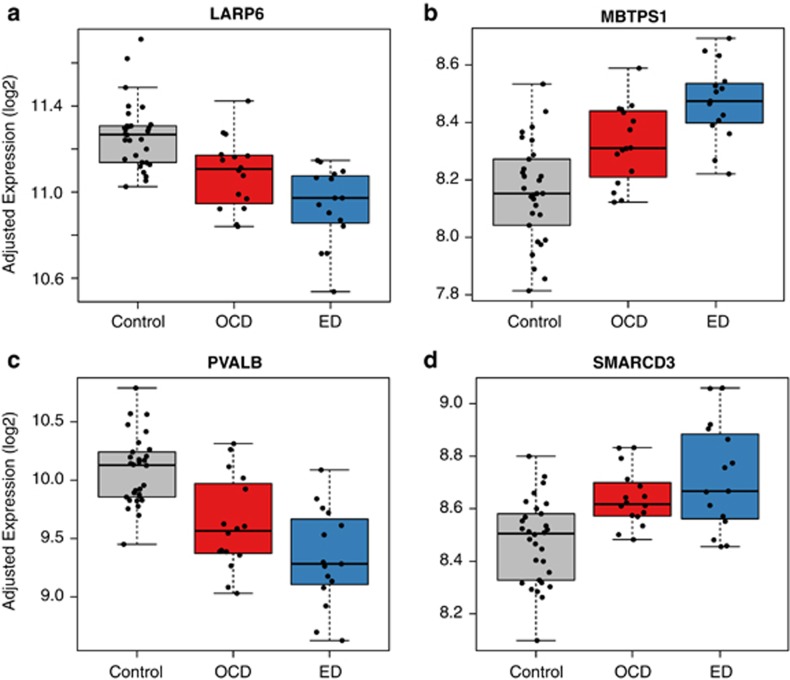
Six genes were differentially expressed comparing ED cases with controls at an FDR of 5% (AK1, LARP6, MBTPS1/S1P, PVALB, RFNG and SMARCD3), with an average fold change of 1.3 (range=1.20–1.61, see Supplementary Table 4). Genetic variation in LARP6 (La ribonucleoprotein domain family, member 6) was significant in a recent GWAS of fasting proinsulin levels in nondiabetic European individuals (P=2.4 × 10−10), and was also marginally significant in association with other glucometabolic traits including fasting insulin and insulin resistance, but not associated with type 2 diabetes32 (Figure 2a).
Figure 2.
Significant differentially expressed genes for eating disorder cases compared with controls for (a) LARP6, (b) MBTPS1, (c) PVALB, and (d) SMARCD3. Note that OCD cases have expression levels between controls and ED cases at these genes. Y axis represents log2 expression, adjusted for estimated surrogate variables and sex. ED, eating disorder; OCD, obsessive-compulsive disorder.
MBTPS1 (a.k.a. S1P; membrane-bound transcription factor pepsidase, site 1) acts in the biogenesis of lysosomes, and has a key role in the Mucolipidosis II disorder via cleavage of the GlcNAc-1-phosphotransferase complex in response to cholesterol deprivation33 (Figure 2b). This disorder is marked by loss of lysosome activity, creating excess oligosaccharides, lipids and glycosaminoglycans, and interestingly, delays in cognitive skills.34 PVALB encodes a protein with high affinity for calcium ion-binding similar to calmodulin and troponin C thought to be involved in muscle relaxation35 and a marker for a subset of fast firing GABA neurons that have been implicated in cognitive processing and in the pathophysiology of various psychiatric disorders (Figure 2c). Lastly, SMARCD3 regulates chromatin structure around target genes, and has a role in neuronal progenitor- and neuronal-specific chromatin remodeling complexes36 (Figure 2d).
While only a few genes reached genome-wide significance, gene set analysis on the global distribution of differentially expressed genes (see Materials and Methods) identified additional genes of potential interest. Functionally, gene expression in KEGG pathways related to mitochondrial respiration and electron transport including oxidative phosphorylation was significantly lower in ED cases (P<5 × 10−8). Similarly, there were decreases in the expression of genes previously identified by Aston et al.37 associated with major depression (N=127 genes; P=4.82 × 10−11) and the KEGG pathway for Parkinson's disease (n=92 genes; P=7.35 × 10−9). There were also several genes sets containing predicted microRNA target genes, including miR637, miR661 and miR661 (see Supplementary Table 5).
Many more genes were differentially expressed between OCD/OCPD/Tic cases (N=15) and controls (N=286), although the average fold change was smaller (1.20, range=1.10–1.49). Given the larger number of differentially expressed genes, we performed gene set analysis using predefined gene sets and Wilcox gene set tests on the genome-wide test statistics. There was significant enrichment for the Blalock collection of Alzheimer's genes38 (P=1.74 × 10−24), genes involved in HeLa cell nuclear phosphoproteins (P=6.32 × 10−13), predicted targets of many microRNAs, and like the ED analysis, genes involved in oxidative phosphorylation (P=6.1 × 10−11; see Supplementary Table 5).
Two of the six significant genes differentially expressed in ED were also significant in patients with OCD (PVALB and RFNG). Furthermore, there was global correlation between the test statistics for OCD and ED among the expressed genes (ρ= 0.483, P<2.2 × 10−16). This overlap prompted a secondary analysis exploring genes that were differentially expressed for both ED and OCD (for example, classifying patients with either OCD or ED as a broad obsessive cohort of cases compared with the controls). There were 1459 differentially expressed genes comparing this more general obsessive psychiatric disorder group with controls (at FDR<5%). Although all six differentially expressed ED genes were contained in this set, only 239 differentially expressed OCD genes were significant here (83.6%). The fold changes for genes associated with diagnosis were similar whether the ED and OCD cases were analyzed together, but we obtained increased power by doubling the sample size of the diagnosis group in the combined analysis (Supplementary Figures 1 and 2).
Obsessive psychiatric syndromes clinical risk SNPs do not associate with gene expression comparing cases and controls
We finally asked whether any of the clinical risk variants could explain the gene expression differences comparing obsessive patients with controls. One important caveat with interrogating postmortem tissue of ill subjects is that it is difficult to untangle whether observed differences in gene expression are associated with the development of illness or result from illness-associated epiphenomena. We attempted to disambiguate these relationships by exploring the association between the previously identified clinical risk SNPs for ED and gene expression of differentially expressed genes (specifically genes with an FDR<5% for ED, OCD/OCPD/Tic only, and the overall broad obsessive cohort; N=6, 286 and 1459, respectively) using the full case–control sample (N=133; genotype is not associated with RIN, and therefore not a confounder in these analyses). Given the small sample size of the patients, we retained the 98 SNPs with MAF>10% (dropping 7 SNPs) in the patient population to avoid spurious findings driven by low minor allele frequencies. In our analysis, clinical risk-associated SNPs did not explain the observed differentially expressed genes.
For example, within the genes associated with ED, none of the SNP-expression pairs were significant within controls (of 588 pairs), and only two pairs were perhaps marginally significant within cases—both pairs were trans associations. First, variants near TOX3 (rs1111482, rs11647880 and rs8062936) had positive association (P=7.20 × 10−4, FDR=0.11) between the minor (and risk) allele and gene expression of RFNG—this gene was more highly expressed in patients with ED. This directional consistency may offer additional biological evidence beyond the nonsignificant statistical association, and may reach genome-wide significance in larger samples sizes. A variant within NEUROD1 (rs1801262) was associated with the expression of SMARCD3 (P=7.79 × 10−4, FDR=0.11). However, this SNP-expression pair did not have directional consistency between the clinical and postmortem clinical finding—the minor allele increases the odds of ED, negatively associates with expression of SMARCD3, but the gene expression levels were higher in ED cases compared with controls. There were no significant (or marginally significant) SNP-expression pairs in either the OCD-associated or broad obsessive-associated differentially expressed genes.
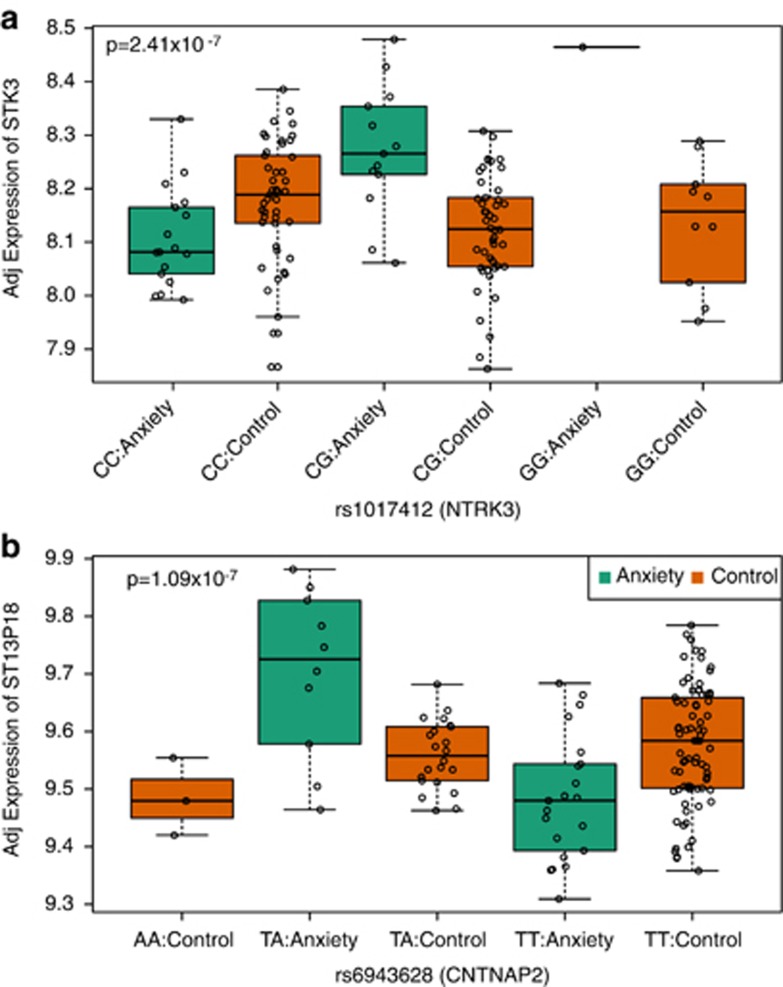
Finally, we reanalyzed all expressed genes (N=12 969) versus the clinical risk variants, and failed to identify any genome-wide significant potential disease-associated (for example, significant interaction between diagnosis, genotype and expression) eQTLs. We do note three marginally significant (each FDR=0.102) SNP-expression pairs associated in trans (Figure 3), as well as slight global enrichment of statistical signal (16 SNP-expression pairs with FDR<20%)—these pairs could possibly reach genome-wide significance in larger studies. The first SNP-expression pair involved genetic variation in NTRK3 (rs1017412), a neurotrophic tyrosine kinase receptor involved in neurotrophin signaling, and the expression levels STK3 (a serine/threonine kinase, P=2.4 × 10−7, FDR=0.155), which have not previously been reported as interacting genes in molecular databases. We also identified potential interaction between genetic variation in CNTNAP2 (rs6943628), a member of the neurexin family and the expression levels of both LOC643310, a pseudogene and TMEM51, a transmembrane protein (P=1.09 × 10−7 and 2.13 × 10−7, respectively).
Figure 3.
Top significant diagnostic eQTLs, identified by significant interaction between genotype and diagnosis for (a) rs1017412 and the expression of STK3 and (b) rs6943628 on the expression of ST13P18. Y axis represents log2 expression, adjusted for estimated surrogate variables. eQTL, expression quantitative trait locus.
Discussion
We identified significant association between a small subset of previously identified genetic risk variants for ED or OCD and gene expression within healthy individuals, and identified hundreds of genes differentially expressed in the DLPFC of patients with both ED and then in a broad obsessive cohort compared with controls. However, we failed to significantly link genetic variation in candidate risk genes with gene expression changes. As none of these risk variants were significantly associated with differentially expressed genes, this perhaps suggests that larger sample sizes may be required to identify both true genome-wide significant risk signals in large clinical GWAS as well as better identify candidate molecular mechanisms of clinical association in postmortem brain data sets. Given that these common risk variants have small effect sizes (odds ratios were between 0.891–1.193 in Boraska et al.12), the small differences in allele frequencies would require hundreds or thousands of samples to potentially identify significant changes in gene expression.
Therefore, the differentially expressed genes for ED may offer biologically relevant targets for future interrogations of genetic risk in large sample sizes and perhaps for untangling pathways related to obsessive symptomatology versus caloric restriction. For example, genetic variation in LARP6, one of the six differentially expressed genes, has previously been implicated in fasting proinsulin levels and marginally significant in other glucometabolic traits including fasting insulin and insulin resistance.32 Abnormalities in these metabolic pathways may have a significant role in ED, such as long-term calorie restriction. Conversely, changes in the expression of S1P and PVALB associated with illness may be more associated with obsessive symptomatology, as both genes have previously been implicated in cognitive skills and processing.34 The gene set analysis also highlights these complementary processes, with lower expression both among genes related to mitochondrial respiration and electron transport including oxidative phosphorylation as well as decreases in the expression of genes associated with major depression and Parkinson's disease.
Although combining ED patients with other more general obsessive psychiatric syndromes may appear controversial, it both granted a much larger sample size to interrogate molecular associations, and also fulfills the research framework proposed in the National Institute of Mental Health's Research Domain Criteria, incorporating common symptoms including obsessionality, perfectionism, anxiety, thought preoccupations, hoarding, a strong need to control one's environment and rigidity that may involve shared genetic risk across these obsessive syndromes. In a recent review of the genetics of ED, Trace et al.39 made a strong case for ‘cross-disorder GWAS', to interrogate the genetics of psychiatric disorders with overlapping features, such as ED and OCD. An important result of our analyses was the overlap between the genes differentially expressed for ED compared with the broad obsessive cohort, suggesting that classifying patients based on meta-categories of observable behavior (that is, obsessionality) may help when searching for the underlying neuropathological changes associated with these related illnesses.
However, the accompanying caveat that many confounds can affect these diagnostic groups in similar fashions may ultimately limit the usefulness of such combined approaches, or at least requires careful matching and statistical analyses. For example, the RNA quality (via RIN) in our patients, although still quite high for human postmortem brain research, was initially lower in cases compared with controls, leadings to thousands of spuriously associated differentially expressed genes. Only after carefully matching for RIN (and thus decreasing our sample size) did we remove these spurious associations. However, other unmeasured confounding factors in a medical examiner sample such as this (for example, death by suicide, acute drug intoxication, psychiatric and substance abuse comorbities and/or prescription medications) may covary with the diagnostic groups, potentially evident in the large increase in number of significant differentially expressed genes when the ED and OCD diagnostic groups were combined. We feel most confident in the differentially expressed genes for ED as the resulting candidate genes had biological plausibility, for example, clinical association with fasting proinsulin levels.32 In addition, the large number of differentially expressed genes comparing patients with OCD with controls may provide a valuable set of candidate genes for more focused clinical associations in larger studies.
The approach used in this study may also be successful in other illnesses for uncovering the molecular mechanisms of clinical risk variants using postmortem brain tissue, namely beginning with established clinical risk variants, identifying allelic variation associated with gene expression in psychiatrically normal individuals, assessing any expression differences associated with diagnosis, and then attempting to link clinical variants with identified illness-related differential expression. The benefit of the first approach, establishing mechanisms of risk in normal individuals, without the confounds of treatment and/or substance abuse, has previously been useful in studies of schizophrenia, Alzheimer's disease and normal human brain development.17,18,20 Here we show that this approach can identify the putative risk gene when clinical risk variants lie in regions with multiple annotated genes.
Although we have identified both significant effects of genetic variation on gene expression, and differentially expressed genes related to diagnosis, the use of more precise measurements tools like RNA sequencing may lead to the identification of particular transcripts of risk that are differentially controlled or expressed in association with illness. We hypothesize that follow-up studies may harness this technology to better untangle the molecular mechanisms of these overlapping disorders. In addition, given that this first-of-its-kind postmortem obsessive cohort is relatively small, given that AN and BN are predominantly manifested in females, and given that many association and neuroimaging studies of ED have been conducted in Caucasian samples, it is imperative that future exploration of risk-associated SNPs in postmortem human control cohorts continue to collect even larger samples, particularly of Caucasian female cases, to increase power and better assess genotype effects. In the case of OCD, additional Caucasian male cases may be of benefit.
This research is the first study to embark on the challenging process of assigning molecular function to genetic clinical risk, motivated by eating and more general obsessive psychiatric disorders, using a human postmortem brain tissue sample.
Data availability
The microarray data, in both raw and processed forms, is available from the Gene Expression Omnibus (GEO) at accession: GSE60190.
Acknowledgments
We gratefully acknowledge the families who donated tissue for this research. We thank the Offices of the Chief Medical Examiner of the District of Columbia and of the Commonwealth of Virginia, Northern District, the National Institute for Child and Health Development Brain and Tissue Bank, the New York State Brain Bank at Columbia University, William E Bunney, MD and Marquis P Vawter PhD at the University of California at Irvine Brain Bank, and the University of Texas Southwest Brain Bank for their collaborative efforts to make this study possible. We also thank Dr Mary Herman, Dr Chao Li and Stefan Difazio for their technical assistance, and we would like to acknowledge the late Mrs Rose G Kleinman. This study was supported by the Intramural Research Program of the National Institute of Mental Health, National Institutes of Health, in Bethesda, MD, USA and the Lieber Institute for Brain Development.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Gorwood P, Bouvard M, Mouren-Simeoni MC, Kipman A, Ades J. Genetics and anorexia nervosa: a review of candidate genes. Psychiatr Genet. 1998;8:1–12. doi: 10.1097/00041444-199800810-00001. [DOI] [PubMed] [Google Scholar]
- Ben-Dor DH, Laufer N, Apter A, Frisch A, Weizman A. Heritability, genetics and association findings in anorexia nervosa. Isr J Psychiatry Relat Sci. 2002;39:262–270. [PubMed] [Google Scholar]
- Stoving RK, Andries A, Brixen K, Flyvbjerg A, Horder K, Frystyk J. Leptin, ghrelin, and endocannabinoids: potential therapeutic targets in anorexia nervosa. J Psychiatr Res. 2009;43:671–679. doi: 10.1016/j.jpsychires.2008.09.007. [DOI] [PubMed] [Google Scholar]
- de Krom M, Bauer F, Collier D, Adan RA, la Fleur SE. Genetic variation and effects on human eating behavior. Annu Rev Nutr. 2009;29:283–304. doi: 10.1146/annurev-nutr-080508-141124. [DOI] [PubMed] [Google Scholar]
- Bergen AW, van den Bree MB, Yeager M, Welch R, Ganjei JK, Haque K, et al. Candidate genes for anorexia nervosa in the 1p33-36 linkage region: serotonin 1D and delta opioid receptor loci exhibit significant association to anorexia nervosa. Mol Psychiatry. 2003;8:397–406. doi: 10.1038/sj.mp.4001318. [DOI] [PubMed] [Google Scholar]
- Bulik CM, Slof-Op't Landt MC, van Furth EF, Sullivan PF. The genetics of anorexia nervosa. Annu Rev Nutr. 2007;27:263–275. doi: 10.1146/annurev.nutr.27.061406.093713. [DOI] [PubMed] [Google Scholar]
- Hinney A, Friedel S, Remschmidt H, Hebebrand J. Genetic risk factors in eating disorders. Am J Pharmacogenomics. 2004;4:209–223. doi: 10.2165/00129785-200404040-00001. [DOI] [PubMed] [Google Scholar]
- Tozzi F, Bulik CM. Candidate genes in eating disorders. Curr Drug Targets CNS Neurol Disord. 2003;2:31–39. doi: 10.2174/1568007033338643. [DOI] [PubMed] [Google Scholar]
- Pinheiro AP, Bulik CM, Thornton LM, Sullivan PF, Root TL, Bloss CS, et al. Association study of 182 candidate genes in anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1070–1080. doi: 10.1002/ajmg.b.31082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root TL, Szatkiewicz JP, Jonassaint CR, Thornton LM, Pinheiro AP, Strober M, et al. Association of candidate genes with phenotypic traits relevant to anorexia nervosa. Eur Eat Disord Rev. 2011;19:487–493. doi: 10.1002/erv.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry. 2011;16:949–959. doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- Boraska V, Franklin CS, Floyd JAB, Thornton LM, Huckins LM, Southam L, et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry. advanced online publication, 11 February 2014. [DOI] [PMC free article] [PubMed]
- American Psychiatric Association . Diagnostic and Statistical Manual for Mental Disorders. American Psychiatric Press: Washington DC, USA; 2000. [Google Scholar]
- Kaye WH, Bulik CM, Thornton L, Barbarich N, Masters K. Comorbidity of anxiety disorders with anorexia and bulimia nervosa. Am J Psychiatry. 2004;161:2215–2221. doi: 10.1176/appi.ajp.161.12.2215. [DOI] [PubMed] [Google Scholar]
- Swinbourne JM, Touyz SW. The co-morbidity of eating disorders and anxiety disorders: a review. Eur Eat Disord Rev. 2007;15:253–274. doi: 10.1002/erv.784. [DOI] [PubMed] [Google Scholar]
- Huffaker SJ, Chen J, Nicodemus KK, Sambataro F, Yang F, Mattay V, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15:509–518. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman JE, Law AJ, Lipska BK, Hyde TM, Ellis JK, Harrison PJ, et al. Genetic neuropathology of schizophrenia: new approaches to an old question, and new uses for post mortem human brains. Biol Psychiatry. 2011;69:140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, et al. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Deep-Soboslay A, Akil M, Martin CE, Bigelow LB, Herman MM, Hyde TM, et al. Reliability of psychiatric diagnosis in postmortem research. Biol Psychiatry. 2005;57:96–101. doi: 10.1016/j.biopsych.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 2013;18:788–798. doi: 10.1038/mp.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortenhorst MS, Wissing MD, Rodriguez R, Kachhap SK, Jans JJ, Van der Groep P, et al. Analysis of the genomic response of human prostate cancer cells to histone deacetylase inhibitors. Epigenetics. 2013;8:907–920. doi: 10.4161/epi.25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos M, Costas J, de Cid R, Bayes M, Gonzalez JR, Baca-Garcia E, et al. Identification of new putative susceptibility genes for several psychiatric disorders by association analysis of regulatory and non-synonymous SNPs of 306 genes involved in neurotransmission and neurodevelopment. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:808–816. doi: 10.1002/ajmg.b.30902. [DOI] [PubMed] [Google Scholar]
- Brown KM, Bujac SR, Mann ET, Campbell DA, Stubbins MJ, Blundell JE. Further evidence of association of OPRD1 & HTR1D polymorphisms with susceptibility to anorexia nervosa. Biol Psychiatry. 2007;61:367–373. doi: 10.1016/j.biopsych.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Meijer H, de Graaff E, Merckx DM, Jongbloed RJ, de Die-Smulders CE, Engelen JJ, et al. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet. 1994;3:615–620. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner K, Kollmann K, Schweizer M, Braulke T, Pohl S. A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science. 2011;333:87–90. doi: 10.1126/science.1205677. [DOI] [PubMed] [Google Scholar]
- Plante M, Claveau S, Lepage P, Lavoie EM, Brunet S, Roquis D, et al. Mucolipidosis II: a single causal mutation in the N-acetylglucosamine-1-phosphotransferase gene (GNPTAB) in a French Canadian founder population. Clin Genet. 2008;73:236–244. doi: 10.1111/j.1399-0004.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- Wahr PA, Michele DE, Metzger JM. Parvalbumin gene transfer corrects diastolic dysfunction in diseased cardiac myocytes. Proc Natl Acad Sci USA. 1999;96:11982–11985. doi: 10.1073/pnas.96.21.11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trace SE, Baker JH, Penas-Lledo E, Bulik CM. The genetics of eating disorders. Annu Rev Clin Psychol. 2013;9:589–620. doi: 10.1146/annurev-clinpsy-050212-185546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data, in both raw and processed forms, is available from the Gene Expression Omnibus (GEO) at accession: GSE60190.