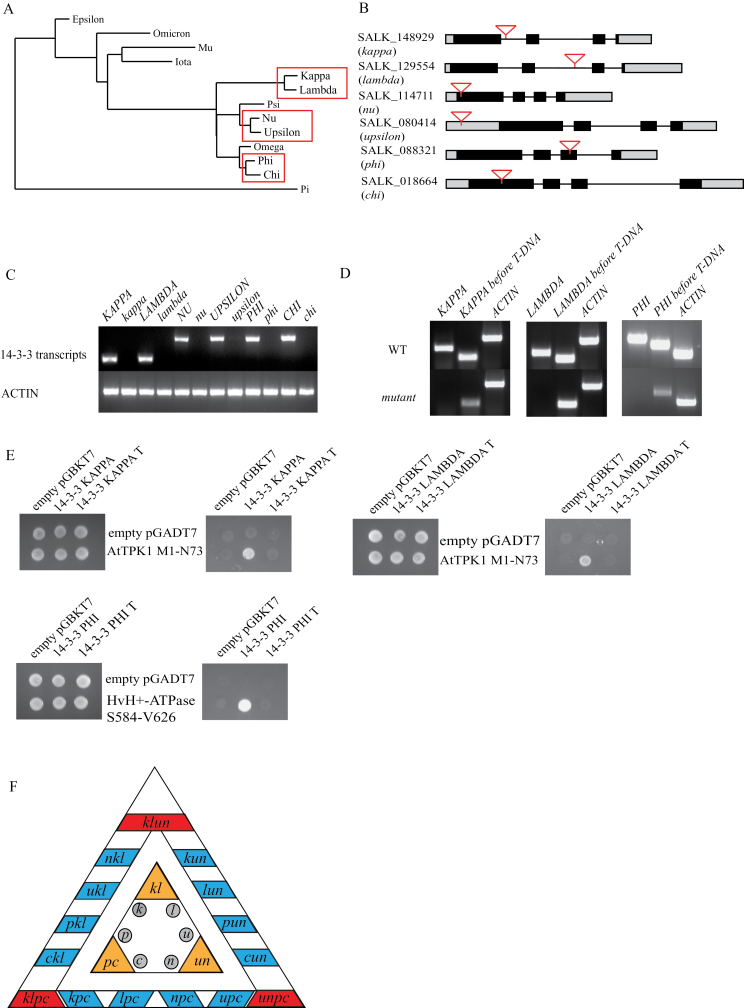
Fig. 1.
14-3-3 T-DNA insertion mutants. (A) Phylogenetic tree of Arabidopsis 14-3-3s with the three closely related gene pairs of which T-DNA insertion lines are used in this study shown in boxes. (B) Position of T-DNA insertion within the six Arabidopsis 14-3-3 genes used in this study. Grey boxes represent UTRs, black boxes exons, lines represent introns, and inverted triangles indicate T-DNA insertion. (C) Expression of 14-3-3 in insertion mutants. Total RNA was isolated from 14 day after stratification (DAS) plants and RT-PCR was used to amplify 14-3-3 transcripts. No full length 14-3-3 transcripts are found in the single mutants. (D) In-frame truncated transcripts have been found in the single mutants kappa, lambda, and phi. (E) Yeast-2-hybrid (Y2H) assay using wild-type 14-3-3s and truncated 14-3-3s (14-3-3 T) showing that the truncated 14-3-3s do not bind to 14-3-3 target proteins, whereas wild-type 14-3-3 does. (F) Overview of the mutants used and generated during this study. The inner triangle depicts the single mutants in grey and at the corners in orange are the double mutants. The outer triangle shows at the sides in blue 12 triple mutants and at the corners in red the quadruple mutants.