Abstract
Despite of the variety of combined modality treatments for laryngeal carcinoma have been introduced, the distance recurrence rate and 5-year overall survival rate over the past decades are still the major issues, underlining the importance to better understand the biological bases that contribute to disease progression. Here, we reported that miR-423-3p overexpressed in primary laryngeal carcinoma cell line where it plays a critical role in tumor progression. Suppression of miR-423-3p expression resulted in decreasing cell proliferation, clonogenicity, cell migration and invasion. By using in silico prediction algorithms for target identification, AdipoR2 (adiponectin receptor 2) and DUSP4 (MAP kinase phosphatase 2) were identified to be potential targets of miR-423-3p. Overexpression of miR-423-3p was associated with epigenetic silencing of AdipoR2 in human laryngeal carcinoma samples, which have been previously implicated in suppression of tumor proliferation and angiogenesis. Luciferase reporter assays and western blot further confirmed the direct interaction of miR-423-3p with AdipoR2. Our findings have demonstrated that miR-423-3p plays an important oncogenic role in laryngeal carcinoma progression, and further suggest that suppression of miR-423-3p expression might be useful for its clinical management.
Keywords: Laryngeal carcinoma, microRNA, target identification
Introduction
Head and neck squamous cell carcinoma is the sixth most common malignant tumour worldwide [1]. Among of which, squamous cell carcinoma of larynx contributes to the second most common type [2]. Smoking and alcohol consumption are the major risk factors and the more commonly affected group is males and aged over 40 years [3]. Because larynx has many essential functions for speech, swallowing and airway protection, the challenge of the treatment is to provide best possible oncologic control, while optimizing functional outcomes. In recent decades, the treatment paradigm for laryngeal cancer, particular for the advanced disease has shifted from primary surgery, toward non-surgical organ-preserving treatment using radiotherapy or chemoradiotherapy [3]. However, despite of the variety of combined modality approaches that have been introduced, the increased local recurrence rate and a worried decreased 5-year overall survival (OS) rates from 48% to 54% have been observed over the past decades [4-6], underlining the importance to better understand the mechanisms that contribute to disease progression and developing novel therapeutic strategies.
MicroRNAs (miRNAs) are a subclass of endogenous non-coding RNAs, which play an essential role in the negatively regulation of gene expression [7]. miRNAs have a pivotal role to regulate a number of biological processes, including development, differentiation, apoptosis and cancer [8]. Recent studies indicate miRNAs can function either as oncogenes or tumor suppressors and have unique ability to regulate many protein-coding genes [9]. Approximately one third of the human genome is regulated by miRNAs, tumor cells exhibit abnormal miRNA expression profiles compared with their normal counterparts [10], suggesting possible role for miRNAs in tumor formation and disease progression. miRNAs in human cancer have been thoroughly investigated. Extensive evidence suggests that some miRNAs may act as selective tools for high-risk patients or miRNAs themselves can be considered as therapeutic targets [8]. However, the biological significance of miRNAs remains largely unknown. Herein, we reported that overexpression of miR-423-3p in human laryngeal cancer cells and identified AdipoR2 and DUSP4 could be potential targets, which may play important roles in laryngeal carcinoma biology. Furthermore, silencing of miR-423-3p led to significant reduction of cell toxicity, suppressed tumor cell migration and invasion in vitro, which in turn, increased AdipoR2 expression, repressed the anti-tumor effects of Adiponectin on the growth of laryngeal cancer, highlighting the biological importance of miR-423-3p in laryngeal cancer progression.
Materials and methods
Cells lines and reagents
Hep-2, the human laryngeal cancer cell line was purchased from the China Center for Type Culture Collection and maintained at 37°C, 5% CO2 in RPMI 1640 complete medium (Gibco, USA) with 10% fetal bovine serum (FBS, Gibco, USA). The normal human hypopharyngeal cells (NHPs) were purchased commercially and cultured in the recommended medium (Celprogen. Inc.). All cells were maintained in a 37°C incubator with humidified 5% CO2. Paired biopsy samples were obtained from five patients with locally advanced laryngeal carcinoma and adjacent normal. Half samples were snap-frozen and stored at -80°C; another half were immediately fixed in 10% buffered-formalin for paraffin embedded. This study was approval from the Second Hospital of Jilin University.
Cell transfections
LipofectAMINE 2000 (Invitrogen) was used to transfect the tumor cells with either scramble (SC) negative control #1 or antimiR-423-3p (life technologies) at a final concentration of 40 nmol/L by using the reverse transfection protocol, according to the manufacturer’s instructions.
Quantification of miRNAs and mRNAs expression
Total RNA was extracted from either cell lines, or primary tumor tissues using RecoverAll Total Nucleic Acid Isolation kit (Ambion). miRNA expression was detected using Taqman MicroRNA Assay (Applied Biosystems). Briefly, the RNA was reverse-transcribed with a MultiScribe reverse transcriptase by using a stem-loop RT primer specifically hybridizes with a miRNA molecule, the RT products were then subsequently amplified with sequence-specific primers using the Applied Biosystems 7900 HT Real-Time PCR system. RNU48 was used as an endogenous control. For mRNAs expressions, the reverse transcription was performed using SuperScript III Reverse Transcriptase (Invitrogen Corp.) according to the manufacturer’s recommendations. qRT-PCR analyses were performed using SYBR Green Master Mix (Applied Biosystems) and the ABI PRISM 7900 Sequence Detection System (Applied Biosystems Inc., Foster City, CA, USA). The relative fold change in mRNA expression was calculated using the 2-ΔΔCt method, where the average of ΔCt values for the amplicon of interest was normalized to that of an endogenous gene (GAPDH), compared with control specimens.
Cell viability and clonogenic assays
The cell proliferation and cytotoxicity of introducing antagomiR, antimiR-423-3p to Hep-2 cells were assessed by using a 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) staining kit (Sigma, USA) [11]. Cells were reverse transfected with either SC control #1, antimiR-423-3p or Lipofectamine 2000 and seeded onto 96-well plates (5 × 103 cells/per well). Cell viability was measured at 24, 48 and 72 hours post-transfection as described previously [guan 2013].
For colony formation assays, transfected Hep-2 cells were seeded into 12-well plates. 48 hours later, cells were harvested, and 500 cells per milliliter were reseeded into 6-well plates in triplicate. After 11 days’ incubation, the plates were fixed and stained, and the number of colonies was then counted. The fraction of surviving cells was calculated by comparing with cells treated with SC.
Cell migration and invasion assays
BD BioCoat Matrigel Invasion Chambers and Control Inserts (BD Biosciences) were used to assess cell migration and invasion. The cells were transfected with either antimiR-423-3p or SC (40 nM), then seeded on either control inserts (polyethylene terephthalate membrane) or trans-well chambers with Matrigel. Two ml RPMI supplemented with 15% FBS was added to the lower chamber, served as the chemo-attractant. 0.5 × 104 transfected cells were re-suspended in RPMI plus 1% FBS, added to the upper chamber (0.5 mls). Twenty hours later, migrating or invading cells attached to the lower surface of the membrane insert were fixed and stained, then counted under a microscope. Relative migration was calculated by comparison with cells transfected with the negative control. The percentage invasion was calculated based on the number of cells which have invaded through the Matrigel insert, divided by the number of cells which have migrated through the control insert.
Immunohistochemistry and Western blotting
Immunohistochemistry was performed using a rabbit anti-AdipoR2 antibody (HPA043737; 1:10 dilution; Sigma-Aldrich) and evaluated using the Level-2 Ultra Streptavidin System (Signet Laboratories) with microwave antigen retrieval. For western blot, the cells were transfected with either SC or antimiR-423-3p. 72 hours post-transfection, the cells were collected and lysed. As described previously [12], protein extracts were prepared and quantified using the BCA method. With 20 μg of protein per well, 15% SDS-PAGE was performed, and samples were then transferred onto Hybond-C membranes (Amersham, USA). Membranes were blocked in 5% milk in Tris-buffered saline with 0.1% Tween-20 (TBST). Blots were incubated with anti-AdipoR2 antibody (1:250) for overnight followed by incubation with the second antibodies (Abcam, USA) labeled with horseradish peroxidase for 2 hours. Signals were visualized using the ECL Western blotting substrate kit (Pierce, USA).
Luciferase reporter assay
The 3’ UTR region of AdipoR2 containing the predicted miR-423-3p binding site was amplified using AmpliTaq gold DNA polymerase (Applied Biosystems). The PCR product was purified, digested with SpeI and HindIII, then cloned downstream of the firefly luciferase gene in the pMIR-REPORT vector (Ambion, Inc.). A mutant sequence was also cloned as a validation plasmid. Either pMirluciferase or pMir-luciferase-gene specific vectors were cotransfected with antimiR-423-3p or SC in Hep-2 cells. pRL-SV vector (Promega) containing Renilla luciferase was also transfected with each condition as a reference control. At 48 hours post-transfection, luciferase activity of Firefly and Renilla luciferase activities were then measured using the Dual-Luciferase Reporter Assay (Promega).
Statistical analysis
All data was expressed as the mean ± SD; a P-value of less than 0.05 was considered to be statistically significant. Statistical Analysis and graphs were completed using Microsoft excel and Graphpad Prism Software (Graphpad Software, Inc.).
Results
miR-423-3p was over expressed in Hep-2 cell line
The expression of miR-423-3p was assessed in human laryngeal carcinoma Hep-2 cells. By comparing with normal NHPs, miR-423-3p significantly up-regulated in Hep-2 cells (Figure 1A). Two fold increasing of miR-423-3p expression were observed, consistent with a previously study [10]. Introduction of anti-miR inhibitor, antimiR-423-3p caused significant reduction of miR-423-3p expression (Figure 1B).
Figure 1.
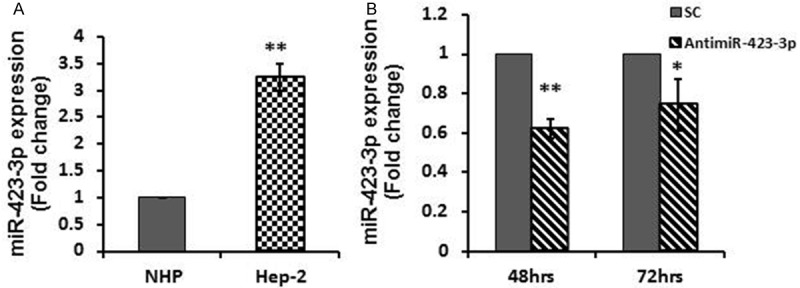
miR-423-3p expression in Hep-2 cell line. A. miR-423-3p expression was detected by real-time RT-PCR. The results showed that expression level of miR-423-3p significantly increased in Hep-2 cell line, compared with normal hypopharyngeal cells (NHPs). B. Depletion of miR-423-3p by utilizing antimiR-423-3p caused significant reduction of miR-423-3p expression in Hep-2 cell line at 48 and 72 hours post-transfection. *P < 0.05; **P < 0.01.
Depletion of miR-423-3p caused significantly reduction of cell viability and decreasing of clonogenicity
The biological significance of depletion of miR-423-3p was analyzed. Hep-2 cells were transfected with 40 nmol/L scramble controls (SC) or antimiR-423-3p. Transfection with antimiR-423-3p into Hep-2 cells led to significantly decreased cell viability compared to controls starting from 24 hours post-transfection, continuously declined at 48 (60%) hours and 72 (58%) hours post-transfection (Figure 2A). Depletion of miR-423-3p also resulted in significant reduction in clonogenicity in Hep-2 cells, colonies formation ratio was 62% compared to 100% of SC (Figure 2B).
Figure 2.
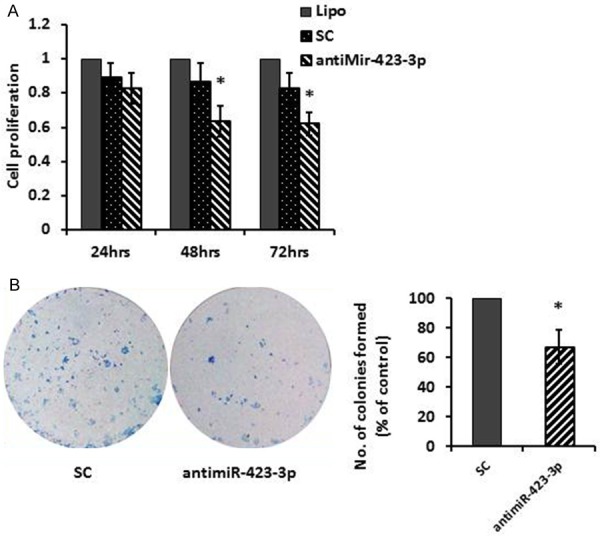
Suppression of miR-423-3p by transfection of antimiR-423-3p reduced cell viability and decreased clonogenic survival. A. Hep-2 cells were transfected with scramble control (SC) antimiR-423-3p (40 nM). Cell viability was measured by the MTT assay 24-72 hours post-transfection. B. Clonogenic survival of Hep-2 cells was assessed 11 days after re-seeding cells treated with SC or antimiR-423-3p (40 nM). *P < 0.05.
miR-423-3p over-expression enhanced cell migration and invasion
To determine whether miR-423-3p involve in regulating cell migration and invasion, in vitro trans-well migration and invasive assays were performed. Compared with their corresponding negative controls, transfection with antimiR-423-3p significantly reduced migrate abilities of Hep-2 cells by 40% and resulted in reduction of invasion of Hep-2 cells by 30%, as shown in Figure 3A and 3B.
Figure 3.
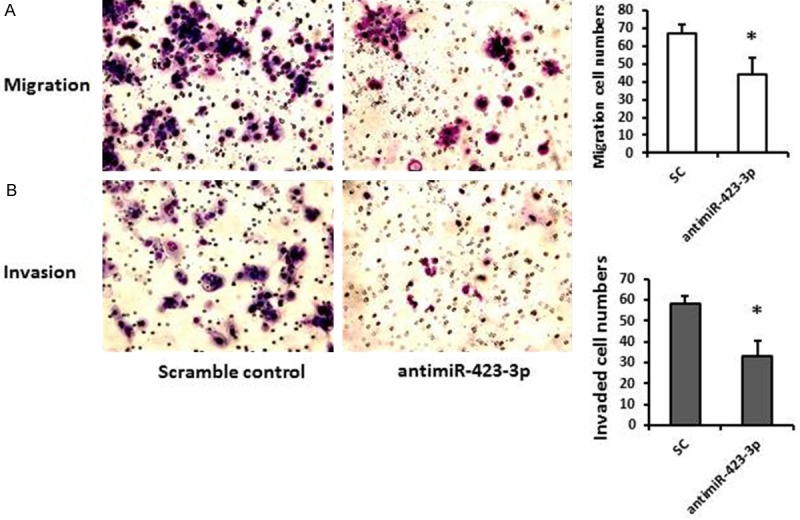
Depletion of miR-423-3p reduced cell migration and invasive. A. Representative images of migration assay indicated that the reduced ability of Hep-2 cells to migrate after transfection with antimiR-423-3p compared to scrambled control (40 nM). B. Representative images of invasive assay indicated that the reduced ability of Hep-2 cells to invade after transfection with antimiR-423-3p compared to scrambled control (40 nM). *P < 0.05.
Identification of miR-423-3p directly mRNA targets
The mechanism of miR-423-3p upregulation in laryngeal carcinoma remains unclear. Here, we used 6 miRNA target-prediction softwares--miRanda, miRDB, miRWalk, PICTAR5, RNA22 and TargetScan to predict potential miR-423-3p targets. 253 mRNAs have interactions with miR-423-3p and were identified as potential targets. Based on their various biological effects on cellular processes, two potential tumor suppressor mRNAs were selected to further study, AdipoR2, adiponectin receptor 2 and DUSP4 (MKP-2), as shown in Figure 4A.
Figure 4.
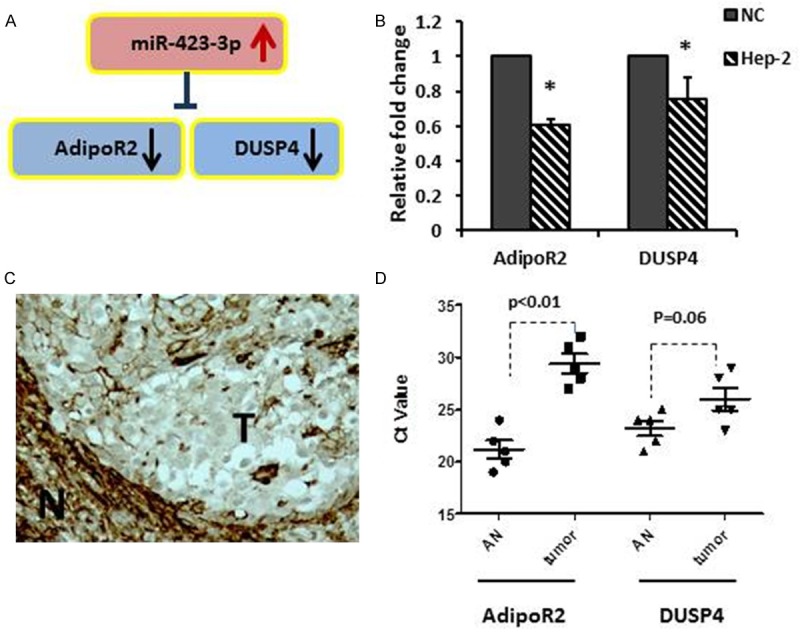
miR-423-3p potential targets identification. A. Identification of two down-regulated genes as potential targets of miR-423-3p. B. qRT-PCR assessment of AdipoR2 and DUSP4 expression in Hep-2 cell lines. *p < 0.05. C. Immunohistochemistry staining showed lower cytoplasmic immunostaining for AdipoR2 in a human formalin-fixed paraffin-embedded laryngeal carcinoma (T), comparing with surrounding normal (N). D. AdipoR2 and DUSP4 expression in 5 paired fresh frozen laryngeal carcinomas (tumor) vs. adjacent normal (AN).
qRT-PCR analyses used to assess the basal expression of AdipoR2 and DUSP4 in Hep-2 cells (Figure 4B). Both AdipoR2 and DUSP4 were significant down-regulated in the cancer cells, compared with normal control (NC). Immunohistochemistry staining showed weak cytoplasmic staining in tumor cells than the surrounding normal cells on 4 out of 5 samples, a representative sample has been shown in Figure 4C. Moreover, AdipoR2 and DUSP4 expression were analyzed in paired laryngeal carcinomas and adjacent normal samples. As showed in Figure 4D, AdipoR2 was significantly down regulated in cancers than in normal, with higher Ct value (P = 0.008). However, DUSP4 showed a border line expression changes between cancer and normal (P = 0.06).
Next, to determine whether miR-423-3p regulates AdipoR2 and DUSP4 by binding to its 3’ UTR, we constructed several reporter vectors carrying the predicted binding site(s) downstream of a firefly luciferase gene in the pMIR-Report vector as previously described [13]. For the luciferase assay, Hep-2 cells were co-transfected with antimiR-423-3p (or a scramble control) and pmiR-AdipoR2-3’ UTR (or a mutated pmiR-AdipoR2-3’ UTR). The luciferase reporter that contained the AdipoR2 3’ UTR was significantly increased by antimiR-423-3p, whereas the mutated reporter was not affected (Figure 5A and 5B). However, we did not observe significant luciferase reporter changes by using pmiR-DUSP4-3’ UTR (or a mutated pmiR-DUSP4-3’ UTR) (Figure 5B). Finally, we further confirmed that the directly interaction between miR-423-3p and AdipoR2 by Western blotting. Overexpression of miR-423-3p (scramble control transfection) led to reduction in AdipoR2 expression at the protein level, while depletion of miR-423-3p (antimiR-423-3p transfection) resulted in significant increasing AdipoR2 expression (Figure 5C).
Figure 5.
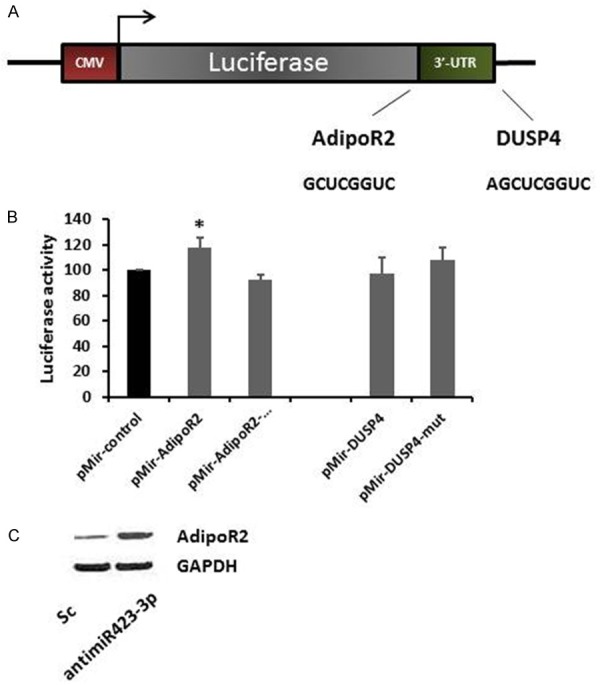
miR-423-3p target validations. A. Illustration of the construction of pmiR-report-AdipoR2 UTR and -DUSP4 UTR vectors. B. Relative luciferase activity after co-transfection with either pmiR-AdipoR2 UTR/pmiR-pmiR-AdipoR2 Mutant vectors or pmiR-DUSP4 UTR/pmiR-DUSP4 UTR Mutant vectors (100 ng for each vectors); scramble and antimiR-423-3p (40 nM), 48 hours post-transfection. Both experiments were performed on Hep-2 cells. C. Western blotting of AdipoR2 in Hep-2 cells was determined 72 hours post-transfection.
Discussion
Extensive studies indicate that miRNAs play important roles in tumor initialization and progression, particular through the regulation of their respective target genes, leading to overexpression of tumor-promoting genes, or down-regulation of tumor-suppressing genes, thereby driving tumor progression [9,13,14]. Herein, we report for the first time that miR-423-3p was significantly up-regulated in Hep-2 laryngeal cancer cells, which promoted tumour cell proliferation, migration and invasion. Furthermore, AdipoR2 (adiponectin receptor 2) was identified as a direct target of miR-423-3p. Overexpression of miR-423-3p was associated with epigenetic silencing of AdipoR2 in Hep-2 cells and human laryngeal carcinoma samples, which in turn led to relieve the suppressive effects of Adiponectin on the growth of laryngeal cancer cells.
miR-423 is localized on chromosome 17q11, a commonly amplified region in breast cancer. Several genes (e.g. TNFAIP1/POLDIP2) in this region strongly correlate with expression of ERBB2 recurrent amplification [15]. miR-423 stays within the first intron of gene NSRP1 (nuclear speckle splicing regulatory protein 1), which is associated with renal cell carcinoma [16]. Pre-mature423 contains two mature transcripts, miR-423-3p and miR-423-5p [17]. miR-423-3p was first described as a mature miRNA in 2004 in untreated HL-60 leukaemia cells following a induced differentiation experiments [18]. Alterations of miR-423 have been reported in multiple cancer types, including hepatocellular carcinoma, head and neck cancer, gastric cancer, colorectal cancer and breast cancer [10,19-22]. However, the exact mechanistic roles of miR-423 in carcinogenesis remain unknown. miR-423 has been reported up-regulation in hepatocellular carcinoma, but only miR-423-3p contributes to the promotion of cell growth and cell cycle progression [19], particular at the G(1)/S transition in hepatocellular carcinoma cells [23]. p21Cip1/Waf1 has been identified as a downstream target of miR-423. Therefore, over-expression of miR-423 promotes hepatic carcinogenesis through the suppression of tumor suppressor p21Cip1/Waf1 expression [23].
Studies have indicated that single nucleotide polymorphisms (SNPs) in miRNAs might promote carcinogenesis by affecting miRNA function and are associated with the development of multiple diseases, including cancer [24,25]. The rs6505162 SNP, an A > C polymorphism located in the pre-miRNA region of miR-423, was found to be associated with a significant overall risk of breast cancer [25]. Compared with the homozygous wild-type genotype, the rs6505162 SNP variants were significantly associated with both overall survival and recurrence-free survival in colorectal cancer [22]. SNP variants in miR-423 further increased the risk for developing OSCC together with environmental smoke [26]. Hence, it is certainly possible that several different mechanisms can account for elevated miR-423 expression in cancer. In our study miR-423-3p appears to play an oncogenic role in laryngeal cancer. Silencing of miR-423-3p significant reduced tumour cell proliferation (Figure 2A), clonogenicity (Figure 2B), cell migration and invasion (Figure 3A and 3B), consistent with other reports in human malignancies [23,27]. The novel finding in our study is that AdipoR2 was identified as a target for miR-423-3p (Figures 4B, 5B, 5C). Adiponectin is an adipocyte-derived cytokine that plays an important role not only in lipid and glucose metabolism but also in the progression of cancer. It has been shown that adiponectin may have anti-cancerous effects by suppressing tumor proliferation and promoting apoptosis [28]. Recent studies demonstrated the antiangiogenic and tumor growth inhibiting properties of adiponectin [29].
Adiponectin binds to two major receptors, AdipoR1 and AdipoR2. Ensuing intracellular signalling pathways link adiponectin with carcinogenesis, with the effect of stimulating AMP-activated protein kinase (AMPK), nuclear factor-κB (NF-κB), peroxisome proliferators-activated receptor (PPAR)-α and mammalian target of rapamycin (mTOR) [30]. Accumulating evidence indicates that adiponectin measurements may serve as a useful prognostic screening tool for early detection of obesity related cancers. The expression of adiponectin receptors in tumor tissues has also been elucidated. AdipoR1 and AdipoR2 were downregulated in human gastric cancer, endometrial adenocarcinoma [29,31]. Knockdown of AdipoR1 and AdipoR2 relieved the suppressive effects of adiponectin on the growth of colon cancer cells [32]. Furthermore, expression of AdipoR2 was inversely associated with T category in oesophageal cancer [33]. Here, we demonstrate decreased transcriptional and protein expression of AdipoR2 in laryngeal cancer cells as well as in archival human laryngeal cancer tissues (Figure 4B-D), providing a translational corroboration between miR-423-3p and AdipoR2. Moreover, we observed an inverse relationship between miR-423-3p and AdipoR2 activation, whereby suppression of miR-423-3p led to an increase in AdipoR2 luciferase activity and protein expression (Figure 5B and 5C). These findings illustrate the ability of the miR-423-3p to interact with AdipoR2 directly.
In summary, we have shown that miR-423-3p plays a novel oncogenic role in laryngeal cancer through regulation of at least one of the adiponectin receptors, AdipoR2, whereby higher miR-423-3p suppresses AdipoR2, in turn relieving the suppressive effects of adiponectin on the tumor growth, leading to increased laryngeal cell proliferation, invasion and migration. These findings suggest an alternate mechanism for AdipoR2, in laryngeal cancer development and progression.
Disclosure of conflict of interest
None.
References
- 1.Wong TS, Gao W, Li ZH, Chan JY, Ho WK. Epigenetic dysregulation in laryngeal squamous cell carcinoma. J Oncol. 2012;2012:739461. doi: 10.1155/2012/739461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Anantharaman D, Marron M, Lagiou P, Samoli E, Ahrens W, Pohlabeln H, Slamova A, Schejbalova M, Merletti F, Richiardi L, Kjaerheim K, Castellsague X, Agudo A, Talamini R, Barzan L, Macfarlane TV, Tickle M, Simonato L, Canova C, Conway DI, McKinney PA, Thomson P, Znaor A, Healy CM, McCartan BE, Hashibe M, Brennan P, Macfarlane GJ. Population attributable risk of tobacco and alcohol for upper aerodigestive tract cancer. Oral Oncol. 2011;47:725–731. doi: 10.1016/j.oraloncology.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman HT, Porter K, Karnell LH, Cooper JS, Weber RS, Langer CJ, Ang KK, Gay G, Stewart A, Robinson RA. Laryngeal cancer in the United States: changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116:1–13. doi: 10.1097/01.mlg.0000236095.97947.26. [DOI] [PubMed] [Google Scholar]
- 5.Mantsopoulos K, Psychogios G, Bohr C, Zenk J, Kapsreiter M, Waldfahrer F, Iro H. Primary surgical treatment of T3 glottic carcinoma: long-term results and decision-making aspects. Laryngoscope. 2012;122:2723–2727. doi: 10.1002/lary.23580. [DOI] [PubMed] [Google Scholar]
- 6.Ganly I, Patel SG, Matsuo J, Singh B, Kraus DH, Boyle J, Wong RJ, Lee N, Pfister DG, Shaha AR, Shah JP. Predictors of outcome for advanced-stage supraglottic laryngeal cancer. Head Neck. 2009;31:1489–1495. doi: 10.1002/hed.21113. [DOI] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulrane L, Klinger R, McGee SF, Gallagher WM, O’Connor DP. microRNAs: a new class of breast cancer biomarkers. Expert Rev Mol Diagn. 2014;14:347–363. doi: 10.1586/14737159.2014.901153. [DOI] [PubMed] [Google Scholar]
- 9.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, Ferreira B, Liu CG, Villanueva A, Capella G, Schwartz S Jr, Shiekhattar R, Esteller M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O’Sullivan B, Waldron J, Gullane P, Cummings B, Liu FF. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 11.Wen LJ, Gao LF, Jin CS, Zhang HJ, Ji K, Yang JP, Zhao XJ, Wen MJ, Guan GF. Small interfering RNA survivin and GRIM-19 co-expression salmonella plasmid inhibited the growth of laryngeal cancer cells in vitro and in vivo. Int J Clin Exp Pathol. 2013 Sep 15;6:2071–2081. [PMC free article] [PubMed] [Google Scholar]
- 12.Guan GF, Zhao M, Liu LM, Jin CS, Sun K, Zhang DJ, Yu DJ, Cao HW, Lu YQ, Wen LJ. Salmonella typhimurium mediated delivery of Apoptin in human laryngeal cancer. Int J Med Sci. 2013 Sep 21;10:1639–1648. doi: 10.7150/ijms.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P’ng C, Miller N, McCready D, Fyles A, Liu FF. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926–2937. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 14.Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, O’Sullivan B, Liu FF. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 15.Grinchuk OV, Motakis E, Kuznetsov VA. Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics. 2010;11(Suppl 1):S9. doi: 10.1186/1471-2164-11-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.miRBase Database. www.mirbase.org. http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0001445. Last accessed 27.6.2014.
- 18.Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- 19.Ma Y, Wang R, Zhang J, Li W, Gao C, Liu J, Wang J. Identification of miR-423 and miR-499 Polymorphisms on Affecting the Risk of Hepatocellular Carcinoma in a Large-Scale Population. Genet Test Mol Biomarkers. 2014;18:516–524. doi: 10.1089/gtmb.2013.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott AM, Miller N, Wall D, Martyn LM, Ball G, Sweeney KJ, Kerin MJ. Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS One. 2014;9:e87032. doi: 10.1371/journal.pone.0087032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Wang X, Yang X, Liu Y, Shi Y, Ren J, Guleng B. miRNA423-5p regulates cell proliferation and invasion by targeting trefoil factor 1 in gastric cancer cells. Cancer Lett. 2014;347:98–104. doi: 10.1016/j.canlet.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 22.Xing J, Wan S, Zhou F, Qu F, Li B, Myers RE, Fu X, Palazzo JP, He X, Chen Z, Yang H. Genetic polymorphisms in pre-microRNA genes as prognostic markers of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2012 Jan;21:217–227. doi: 10.1158/1055-9965.EPI-11-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J HS, Wu S, Ding J, Zhao Y, Liang L, Tian Q, Zha R, Zhan R, He X. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011 Nov;32:1641–1647. doi: 10.1093/carcin/bgr199. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Yu CY, Wang JL, Guan J, Chen HY, Fang JY. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci Rep. 2014;4:3648. doi: 10.1038/srep03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith RA, Jedlinski DJ, Gabrovska PN, Weinstein SR, Haupt L, Griffiths LR. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genomics Proteomics. 2012 May-Jun;9:115–118. [PubMed] [Google Scholar]
- 26.Wang Y, Vogelsang M, Schäfer G, Matejcic M, Parker MI. MicroRNA polymorphisms and environmental smoke exposure as risk factors for oesophageal squamous cell carcinoma. PLoS One. 2013;8:e78520. doi: 10.1371/journal.pone.0078520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, Deng T, Liu H, Yin J, Wang S, Zen K, Ba Y, Zhang CY. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011 Mar;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 28.Kelesidis I, Kelesidis T, Mantzoros CS. Adiponectin and cancer: a systematic review. Br J Cancer. 2006;94:1221–1225. doi: 10.1038/sj.bjc.6603051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamauchi N, Takazawa Y, Maeda D, Hibiya T, Tanaka M, Iwabu M, Okada-Iwabu M, Yamauchi T, Kadowaki T, Fukayama M. Expression levels of adiponectin receptors are decreased in human endometrial adenocarcinoma tissues. Int J Gynecol Pathol. 2012;31:352–357. doi: 10.1097/PGP.0b013e3182469583. [DOI] [PubMed] [Google Scholar]
- 30.Dalamaga M, Diakopoulos KN, Mantzoros CS. The role of adiponectin in cancer: a review of current evidence. Endocr Rev. 2012;33:547–594. doi: 10.1210/er.2011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otani K, Kitayama J, Kamei T, Soma D, Miyato H, Yamauchi T, Kadowaki T, Nagawa H. Adiponectin receptors are downregulated in human gastric cancer. J Gastroenterol. 2010;45:918–927. doi: 10.1007/s00535-010-0228-2. [DOI] [PubMed] [Google Scholar]
- 32.Hiyoshi M, Tsuno NH, Otani K, Kawai K, Nishikawa T, Shuno Y, Sasaki K, Hongo K, Kaneko M, Sunami E, Takahashi K, Nagawa H, Kitayama J. Adiponectin receptor 2 is negatively associated with lymph node metastasis of colorectal cancer. Oncol Lett. 2012 Apr 1;3:756–760. doi: 10.3892/ol.2012.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howard JM, Cathcart MC, Healy L, Beddy P, Muldoon C, Pidgeon GP, Reynolds JV. Leptin and adiponectin receptor expression in oesophageal cancer. Br J Surg. 2014;101:643–652. doi: 10.1002/bjs.9469. [DOI] [PubMed] [Google Scholar]


