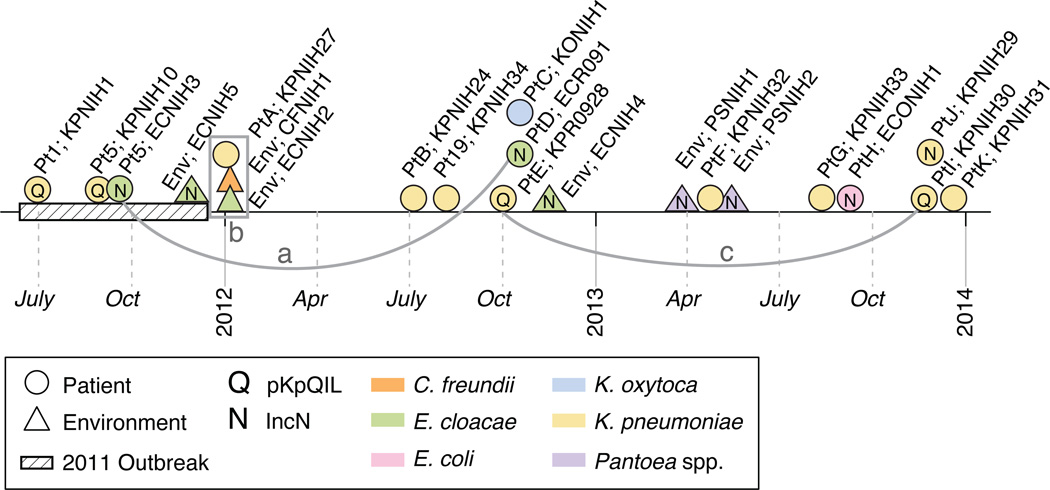
Fig. 1.
Timeline of KPC+ Enterobacteriaceae organisms identified and sequenced from 2011–2013. Patient code, genus, species and strain denoted in label; for example, Pt1; KPNIH1 is Patient 1;Klebsiella pneumoniae NIH1 strain and Env;ECNIH5 is Environmental; Enterobacter cloacae NIH5 strain. Detailed data are presented in Table 1. The isolate source is denoted by shape and the bacterial species is denoted by color. Isolates carrying a variant of the plasmid pKpQIL are marked with a “Q”. Isolates carrying the pKPC-47e backbone (IncN, this study) are marked with an “N”. Isolates connected by lines a and c share genetic similarity, without epidemiologic linkage. Isolates in box b show evidence of an inter-genus exchange of a pR55-like plasmid. The 2011 KPC+ K. pneumoniae outbreak is denoted with a hashed line.