SUMMARY
TRIM58 is an E3 ubiquitin ligase superfamily member implicated by genome wide association studies (GWAS) to regulate human erythrocyte traits. Here we show that Trim58 expression is induced during late erythropoiesis and that its depletion by shRNAs inhibits the maturation of late stage nucleated erythroblasts to anucleate reticulocytes. Imaging flow cytometry studies demonstrate that Trim58 regulates polarization and/or extrusion of erythroblast nuclei. In vitro, Trim58 directly binds and ubiquitinates the intermediate chain of the microtubule motor dynein. In cells, Trim58 stimulates proteasome-dependent degradation of the dynein holoprotein complex. During erythropoiesis, Trim58 expression, dynein loss and enucleation occur concomitantly and all are inhibited by Trim58 shRNAs. Dynein regulates nuclear positioning and microtubule organization, both of which undergo dramatic changes during erythroblast enucleation. Thus, we propose that Trim58 regulates this process by eliminating dynein. Our findings identify an erythroid-specific regulator of enucleation and elucidate a previously unrecognized mechanism for controlling dynein activity.
INTRODUCTION
Humans produce about 2 million red blood cells (erythrocytes) each second to replace those lost by senescence (McGrath and Palis, 2008). Erythropoiesis is accompanied by several specialized cell divisions associated with activation of erythroid-specific genes (Cheng et al., 2009; Welch et al., 2004), repression of alternate lineage genes (Cheng et al., 2009; Kingsley et al., 2013; Welch et al., 2004), global DNA demethylation (Shearstone et al., 2011), reduced cell volume (Dolznig et al., 1995) and ejection of the nucleus to yield a reticulocyte, which develops further into a mature red blood cell (Liu et al., 2010). The mechanisms that govern these processes are incompletely understood.
Expulsion of nuclei from erythroblasts (Keerthivasan et al., 2011; Konstantinidis et al., 2012) occurs exclusively in mammals and may represent an evolutionary adaptation to optimize erythrocyte rheology for transport through small capillary beds (Gaehtgens et al., 1981; Mueller et al., 2008). Erythroblast enucleation is preceded by nuclear condensation and requires histone deacetylation (Ji et al., 2010; Popova et al., 2009) and suppression of the Myc proto-oncogene (Jayapal et al., 2010). The condensed nucleus polarizes to one side of the cell via transport mechanisms that require microtubules and phosphoinositide 3-kinase (Wang et al., 2012), followed by Rac GTPase-mediated formation of a contractile actomyosin ring between the incipient reticulocyte and nucleus (Ji et al., 2008; Konstantinidis et al., 2012). Forces generated by the contractile ring promote further nuclear extrusion (Ji et al., 2008; Wang et al., 2012). Final separation between the reticulocyte and nucleus is facilitated by transport of lipid vesicles to the interface, which facilitates remodeling and resolution of the plasma membrane surrounding both structures (Keerthivasan et al., 2010; Keerthivasan et al., 2011). In vivo, the ejected nucleus with surrounding plasma membrane, termed a pyrenocyte (McGrath et al., 2008), is scavenged by macrophages (Soni et al., 2006; Yoshida et al., 2005), while reticulocytes are released into the circulation and undergo further maturation (Gifford et al., 2006). Enucleation shares several features with cytokinesis (Keerthivasan et al., 2010; Konstantinidis et al., 2012). All of the proteins currently known to participate in erythroblast enucleation are ubiquitously expressed, with functional roles in mitosis. How erythroblasts co-opt generalized mitotic machinery for enucleation is unknown. Presumably, this is mediated in part by the expression of one or more erythroid-restricted proteins.
Here, we identify a role for the erythroid protein Trim58 in erythroblast enucleation. Trim58 is a member of the tripartite motif-containing family of proteins that frequently possess E3 ubiquitin ligase activities and function broadly in physiology and disease (Napolitano and Meroni, 2012). Genome wide association studies (GWAS) show that single nucleotide polymorphisms (SNPs) linked to the human TRIM58 gene associate with variations in the size and/or number of circulating erythrocytes (Kamatani et al., 2010; van der Harst et al., 2012). However, SNPs identified by GWAS do not necessarily reflect the activity of the nearest gene (Sankaran and Orkin, 2013). We performed functional studies to investigate the role of Trim58 in erythropoiesis. Our findings suggest that Trim58 facilitates erythroblast enucleation by inducing proteolytic degradation of the microtubule motor dynein. Thus, we identify a lineage-restricted protein that participates in erythroblast enucleation, likely by targeting a ubiquitous protein complex that is essential for most other eukaryotic cells.
RESULTS
Trim58 is induced during late stage erythropoiesis
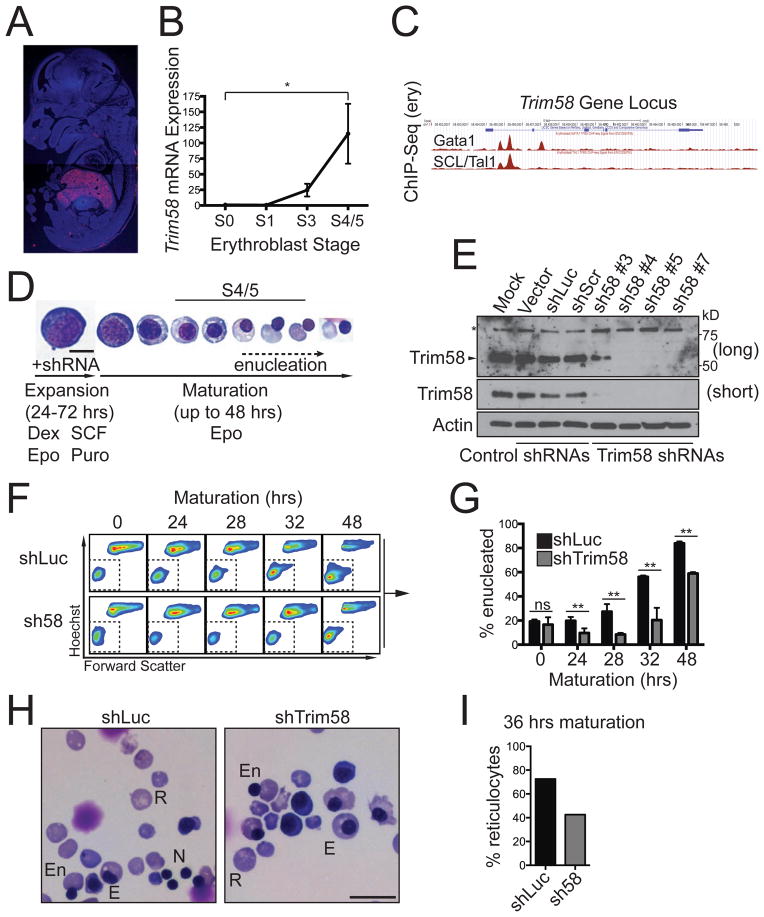
TRIM58 expression is particularly high in the erythroid lineage (Figures S1A and S1B) and is strongly induced during late maturation (Figure S1C). Trim58 mRNA was predominantly expressed in embryonic day 14.5 (E14.5) mouse fetal liver, an erythropoietic tissue (Figure 1A). Real time PCR showed that Trim58 mRNA is upregulated >100-fold in late stage murine fetal liver erythroid precursors (Figure 1B). Chromatin immunoprecipitation-sequencing (ChIP-Seq) of primary murine erythroblasts demonstrated that the essential erythroid transcription factors Gata1 and SCL/Tal1 bind the Trim58 locus within the first intron, a common location for erythroid enhancers (Figure 1C) (Cheng et al., 2009; Pimkin et al., 2014). Thus, Trim58 is strongly and specifically expressed during late erythroid maturation, in part via direct activation by key hematopoietic transcription factors.
Figure 1. Trim58 is expressed during late stage erythropoiesis and regulates erythroid maturation.
(A) RNA in situ hybridization for Trim58 mRNA in an embryonic (E) day 14.5 mouse embryo showing strong expression (red) in fetal liver (FL), the site of definitive erythropoiesis. (B) E14.5 FL erythroblasts were flow cytometry-purified (Pop et al., 2010) and Trim58 mRNA was analyzed by semiquantitative real-time PCR. The y-axis shows relative mRNA expression, normalized to S0 cells, which were assigned a value of 1. The x-axis shows progressive developmental stages from less to more mature. The results represent mean ± SEM for 3 biological replicates. *p<0.05. (C) Chromatin immunoprecipitation-sequence (ChIP-Seq) analysis of transcription factor binding to the Trim58 locus in E14.5 FL erythroblasts (data from (Pimkin et al., 2014)). The blue line depicts the Trim58 gene, with exons shown as rectangles. Transcription factor binding sites are indicated in red. (D) Trim58 knockdown studies. E14.5 murine FL erythroid precursors were purified, infected with retroviruses encoding Trim58 or control shRNAs, and cultured for 24–72 hours in expansion medium with puromycin (Puro) to inhibit terminal maturation and select for infected cells. The cells were then switched to maturation medium, which facilitates development to the reticulocyte stage over ~48 hours. Late stage S4/5 cells are small and hemoglobinized with condensed nuclei (see panel B) (Pop et al., 2010). Epo, erythropoietin; SCF, stem cell factor; Dex, dexamethasone. Scale bar, 10 μm. (E) Western blot for Trim58 in erythroblasts expressing Trim58 or control shRNAs after 48 hours maturation. Luc, luciferase; Scr, scrambled. The asterisk (*) represents a nonspecific band. “Long” and “short” exposures are from the same blot. (F) Enucleation of control (shLuc) and Trim58-deficient (shTrim58 #4) erythroblasts measured by flow cytometry the indicated time points. During maturation, erythroblasts become smaller, indicated by decreased forward scatter, and ultimately enucleate to become Hoechst-negative reticulocytes (boxed regions). (G) Percent (%) enucleation over time in cultures treated with control (shLuc) or Trim58 shRNA #4. Mean ± SD for 3 biological replicates. (H) Representative histology of control (shLuc) and Trim58-deficient (shTrim58 #4) erythroid cultures after 36 hours maturation. Examples of anucleate reticulocytes (R), nucleated erythroblasts (E), extruded nuclei (N) and erythroblasts in the process of enucleation (En) are shown. Scale bar, 20 μm. (I) Summary of reticulocyte fractions from panel H. Six hundred cells were counted on each slide. See also Figures S1 and S2.
The expression pattern of Trim58 contrasts with other E3 ubiquitin ligases that regulate earlier stages of erythroid development, such as Trim10/Herf1 (Harada et al., 1999), Trim28 (Hosoya et al., 2013), Mdm2 (Maetens et al., 2007), and Mdm4 (Maetens et al., 2007) (Figure S1D). Trim58 mRNA was not detected during induced maturation of the murine erythroid cell lines G1E-ER4 (Weiss et al., 1997) or murine erythroleukemia (MEL), likely because they do not mature to late stages (Figure S1E).
Trim58 regulates erythroblast enucleation
We used shRNAs to suppress Trim58 expression during in vitro maturation of primary murine fetal liver erythroblasts (Figure 1D) (Zhang et al., 2003). We infected purified E14.5 erythroid precursors with retroviruses encoding Trim58 or control shRNAs along with green fluorescent protein (GFP) and puromycin resistance cassettes (Hemann et al., 2003), then cultured for 1–3 days with dexamethasone, stem cell factor (SCF), erythropoietin (Epo) and puromycin to promote expansion of infected immature erythroblasts. Shifting to medium containing Epo as the sole cytokine induced terminal maturation. Four different shRNAs reduced Trim58 mRNA and protein by 60–90% (Figures 1E and S2A). During late maturation, erythroblasts expel their nuclei to become anucleate Hoechst− reticulocytes (Figure 1F). The kinetics of enucleation were delayed in Trim58-deficient cultures (Figure 1G) and enucleation was consistently inhibited at 48 hours maturation by all four Trim58 shRNAs compared to controls (Figures 1F, 1G, and S2B). Histological staining confirmed these findings, showing reduced proportions of reticulocytes in Trim58-deficient cultures (Figures 1H and 1I). Trim58 suppression also increased the proportions of mature erythroblasts containing two or more nuclei (Figure S2C).
Numerous parameters of erythroid maturation were not altered by Trim58 knockdown, including downregulation of the cell surface marker CD44 (Figure S2D) (Chen et al., 2009), hemoglobin accumulation (Figure S2E) and nuclear condensation (Figure S2F). Trim58 knockdown produced only small and inconsistent effects on erythroblast proliferation (Figure S2G) and viability (Figure S2H). Overall, these findings demonstrate that Trim58 depletion causes selective defects specifically during late stage erythropoiesis, including reduced enucleation and increased formation of multinucleated cells.
Trim58 binds the molecular motor dynein
Trim58 is predicted to be an E3 ubiquitin ligase with several functional modules, including a PRY-SPRY (PS) domain that mediates substrate interactions (Figure 2A) (James et al., 2007; Woo et al., 2006). We performed pull down studies to identify Trim58-binding proteins, including potential ubiquitination substrates. We used the isolated PS domain for these studies because ectopic expression of full-length wild type (WT) Trim58 was toxic to erythroblasts (data not shown). We expressed FLAG epitope-tagged PS domain in the erythroblast cell line G1E (Weiss et al., 1997), which contains no endogenous Trim58 protein (Figure S1E), immunoprecipitated (IP) with FLAG antibody and analyzed the recovered proteins by SDS-polyacrylamide gel electrophoresis. This analysis identified five discrete protein bands (Figure 2B). Mass spectrometry revealed that these bands contained multiple subunits of the dynein cytoplasmic microtubule motor protein complex (Table 1), as well as nuclear pore complex proteins, a Golgi component, and several other proteins (Table S1). Dynein regulates nuclear positioning and microtubule structure within cells (McKenney et al., 2010; Splinter et al., 2010). Since erythroblast enucleation is microtubule-dependent (Konstantinidis et al., 2012; Wang et al., 2012), we focused on the interaction between Trim58 and dynein.
Figure 2. Trim58 binds directly to the molecular motor dynein.
(A) Modular structure of Trim58 showing the RING, BBox, Coiled Coil (CC) and immunoglobulin-like PRY-SPRY (PS) domains with relevant amino acids numbered. In some Trim proteins, the RING domain recruits E2 conjugases carrying activated Ubiquitin and the PS domain binds substrates. (B) FLAG-tagged Trim58 PS domain or vector (V) were stably expressed in the G1E proerythroblast line. Lysates were immunoprecipitated (IP) with FLAG antibody, fractionated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie Blue Silver. Numbers denote visible bands from PS immunoprecipitates that were excised for mass spectrometric (MS) analysis. The same regions from the control (V) lane were analyzed similarly. MS identified exclusively in the PS lane peptides from dynein heavy chain (DHC), dynein intermediate chain (DIC), dynein light intermediate chains 1/2 (DLIC) and Trim58 PS, as indicated for each band. See also Tables 1 and S1. Inp, input (1%). (C) Lysates from G1E cells expressing FLAG-PS or V were immunoprecipitated with FLAG antibody and analyzed by Western immunoblotting (IB) for DHC, DIC, FLAG and Codanin1 (Cdan1, negative control). Inp, input (5%). (D) E14.5 murine FL erythroblasts were lysed, immunoprecipitated with anti-DIC antibody or IgG control and analyzed by Western blotting for the indicated proteins. α globin represents a negative control. Inp, input (0.5%). (E) Recombinant purified GST-Trim58 PS domain (GST-PS) or GST were incubated with purified bovine holodynein complex and glutathione-Sepharose beads. Bound proteins were analyzed by Western blotting. Equal percentages of total input (Inp), pull down (PD) and supernatant (SN) samples were loaded. (F) 293T cells were transfected with expression plasmids encoding hemagglutinin (HA) fused to the indicated DIC amino acids. After 24 hours, cells were lysed and incubated with GST-PS or GST and glutathione-Sepharose beads. Bound proteins were analyzed by anti-HA Western blotting. PD, pull down; SN, supernatant (1%). (G) Size-exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS) data for PS, GST-DIC (1–120) and a 1:1 mixture. Protein concentration was measured on an inline refractive index detector. Light scattering data converted to molecular weight are shown above each chromatography trace and relate to the right-hand y-axis. The observed molecular weights are consistent with a 1:1 interaction between the two proteins augmented by the dimerization of the GST tag appended to DIC (1–120). (H) DIC domain structure showing the amino-terminal coiled coil (CC) motif, which binds Trim58. aa#, amino acid number of DIC. See also Figure S3.
Table 1. The Trim58 PS domain interacts with multiple dynein subunits.
FLAG-Trim58 PS domain or FLAG alone (Vector Control) was expressed in G1E erythroblasts, purified by FLAG IP and fractionated by SDS-PAGE. Protein bands depicted in Figure 2B were analyzed by mass spectrometry. Trim58 PS-interacting dynein subunits are indicated in this Table and all other proteins are shown in Table S1. MW, molecular weight.
| Protein Name | Accession | MW | Band | Unweighted Spectra | Unique Peptides | Percent Coverage |
|---|---|---|---|---|---|---|
| Cytoplasmic dynein 1 | ||||||
| Heavy chain 1 | IPI00119876 | 532 kD | 1 | 189 | 95 | 19 |
| Intermediate chain 2 | IPI00131086 | 68 kD | 3 | 16 | 7 | 14 |
| Light intermediate chain 1 | IPI00153421 | 57 kD | 3,4 | 38 | 12 | 27 |
| Light intermediate chain 2 | IPI00420806 | 54 kD | 4 | 12 | 7 | 16 |
| Trim58 PRY-SPRY (Bait) | ||||||
| Trim58 | IPI00353647 | 55 kD | 4,5 | 91 | 14 | 26 |
We confirmed the Trim58 PS domain-dynein interaction in G1E cells and transiently transfected 293T cells by FLAG-IP followed by Western blots for dynein intermediate chain (DIC) and dynein heavy chain (DHC) (Figures 2C and S3A). Furthermore, DIC IP from mouse fetal liver recovered Trim58, demonstrating an interaction between the endogenous proteins (Figure 2D). Bacterially expressed GST-tagged Trim58 PS domain captured purified holodynein complex, indicating a direct interaction (Figures 2E and S3B). PS domains from Trim proteins 9, 10, 27 or 36 did not bind dynein, demonstrating specificity for the interaction with Trim58 (Figures S3A and S3B). The dynein multi-subunit complex contains DHC, DIC, light intermediate chains, and light chains. DIC mediates dynein interactions with several accessory proteins, mainly via the amino terminus (McKenney et al., 2011; Vallee et al., 2012). To test whether Trim58 interacts with DIC, we expressed hemagglutinin (HA)-tagged segments of the DIC amino terminus in 293T cells, incubated lysates with GST proteins and glutathione-Sepharose beads, and analyzed interacting polypeptides (Figure 2F). Western blots for HA showed that PS interacted with DIC through an interface within the first 73 amino acids. We used size-exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS) to confirm this interaction in vitro (residues 1–120). Purified bacterially expressed PS and GST-DIC (1–120) each eluted in gel filtration as a single peak when run individually (Figure 2G). The MALLS data, which provide a reference-free estimate of solution molecular weight, showed that PS eluted as a monomer (observed MW = 25.6 kD; expected mass = 23.3 kD), whereas GST-DIC (1–120) ran as a dimer (observed MW = 91.2 kD; expected mass of dimer = 79.6 kD), reflecting the known dimerization of GST (Fabrini et al., 2009). An equimolar mixture of PS and GST-DIC eluted predominantly as a distinct single peak with a lower retention time, indicating formation of a complex. The molecular weight of this signal calculated from MALLS data (126.2 kD) is consistent with a 2:2 complex (expected mass = 114.4 kD) that represented a 1:1 Trim58-DIC interaction that additionally dimerized through the GST moiety. Together, these data show that the Trim58 PS domain binds dynein directly through the DIC amino terminus (Figure 2H). This region of DIC contains a coiled coil domain that interacts with other dynein regulatory proteins, including the dynactin subunit p150Glued and NudE (Ma et al., 1999; McKenney et al., 2011).
Trim58 ubiquitinates dynein and promotes its proteasomal degradation
We expressed FLAG/mCherry-tagged full length WT Trim58 in HeLa cells, which do not express endogenous Trim58, and assessed the effects on dynein protein levels. As controls, we expressed vector alone (V) or a “RING-dead” (RΔ) Trim58 containing two missense mutations predicted to abrogate E3 ligase activity (Figure 3A) (Zhang et al., 2012). The cultures were treated with puromycin for 3 days to enrich for infected cells and analyzed by Western blotting (Figure 3B). Although WT Trim58 slowed cell proliferation, we were able to create viable, stably expressing lines. Compared to the RΔ mutant, WT Trim58 was poorly expressed, possibly due its autoubiquitination and subsequent proteasomal degradation, which occur with other Trim proteins (Versteeg et al., 2013). Dynein subunits DHC and DIC were nearly absent from cells expressing WT Trim58, but not from cells expressing the RΔ version or vector alone (Figure 3B and data not shown). In contrast, the level of DYNC1I2 mRNA was unaffected by WT Trim58 expression despite reduction of the corresponding DIC protein (Figure S4). Treatment with the proteasome inhibitor MG132 partially restored dynein and Trim58 expression (Figure 3B). E. coli-derived recombinant Trim58 exhibited autoubiquitination activity (Figure 3C). In vitro, recombinant WT, but not RΔ, Trim58 ubiquitinated GST-DIC (1–120) (Figure 3D). These data indicate that Trim58 polyubiquitinates dynein to mediate its proteasomal degradation in cells.
Figure 3. Trim58 ubiquitinates dynein and promotes its proteasomal degradation.
(A) Vector (V) and full-length Trim58 constructs with amino-FLAG and carboxyl-mCherry tags. The RΔ mutant contains two missense mutations that abrogate E3 ubiquitin ligase activity. (B) Trim58-expressing HeLa cells were infected with retrovirus encoding FLAG-Trim58 (WT or RΔ) or vector (V), selected in puromycin for 3 days, then analyzed by Western blotting before or after treatment with the proteasome inhibitor MG132 (10 μM) for 4 hours at 37 °C. See also Figure S4. (C) In vitro ubiquitination assay. Recombinant GST-tagged proteins were incubated with E1, E2 (UBE2D3) and HA-tagged Ubiquitin (Ub) for 1 hour at 37 °C, followed by Western blotting for HA or GST. Trim63 was used as a positive control. The high molecular weight smears indicate poly-HA-Ub attachment to the GST fusion proteins. (D) In vitro ubiquitination assay. GST-DIC (1–120) was incubated with WT or RΔ Trim58, E1 and E2 enzymes, and HA-Ub for 1 hour at 37 °C, followed by Western blotting using an anti-DIC antibody. Two arrowheads indicate DIC protein, where the higher one denotes GST-DIC (1–120), and the lower is DIC (1–120). The asterisk (*) represents a nonspecific band. (E) Golgi distribution in HeLa cells expressing Trim58. Cells were transfected with expression plasmids encoding Trim58-mCherry or CC1-mCherry, a dynein inhibitor (Quintyne et al., 1999). After 36 hours, the cells were fixed, stained for the Golgi matrix protein GM130 and DNA (DAPI), and visualized by confocal microscopy. The percentages of mCherry-positive cells that displayed normal punctate perinuclear Golgi body distribution are shown at right as mean ± SD for 3 independent experiments, with >100 cells counted per experiment. **p<0.01 vs. vector control; ns, not significant. (F) Mitotic progression in HeLa cells expressing Trim58-mCherry proteins, as analyzed by time-lapse microscopy. Representative images show mitotic progression in outlined cells. Expression of WT Trim58 delayed progression from cell rounding (~metaphase) to anaphase onset, compared to cells expressing vector or RΔ Trim58. BF, brightfield; mCh, mCherry. Scale bar (upper left panel), 16 μm. (G) Quantitative analysis of mitosis in Trim58-expressing HeLa cells. The graph on the left shows time elapsed between cell rounding and anaphase with mean ± SD for all observed mitotic cells. The percentages of cells with delayed (>200 min) or failed mitosis manifested as cell death between rounding and anaphase are shown on the right. (V, n=134 normal mitoses; WT, n=50 normal, 13 delayed, 11 failed mitoses; RΔ, n=100 normal, 7 delayed mitoses). **p<0.01; ns, not significant. See also Movies S1A–D.
We investigated whether Trim58 expression elicited cellular phenotypes characteristic of dynein loss. Dynein transports Golgi bodies along microtubules into perinuclear stacks at the microtubule organizing center (MTOC) (Quintyne et al., 1999). Inhibition of dynein by expression of CC1, a portion of p150Glued that interferes with the DIC-dynactin interaction, causes Golgi fragmentation and dispersal throughout the cytoplasm (Quintyne et al., 1999). We verified this finding in HeLa cells and showed that WT, but not RΔ, Trim58 produced similar effects (Figure 3E). Dynein also functions in mitotic spindle checkpoint inactivation to facilitate anaphase onset (Howell et al., 2001). Ectopic expression of WT Trim58 in HeLa cells caused mitotic defects characteristic of dynein inhibition, including prolonged interval between cell rounding and anaphase (Figures 3F and 3G and Movies S1A–D) and cell death after rounding (Figure 3G right panel and Movie S1C) that were significantly worse than cells expressing vector or RΔ Trim58. Thus, ectopic expression of Trim58 in heterologous cells induces phenotypes that are consistent with dynein deficiency.
Trim58 expression in erythroblasts coincides with loss of dynein and enucleation
In murine erythroblast cultures undergoing semi-synchronous maturation, upregulation of endogenous Trim58 at 24 hours correlated with loss of dynein subunits (DIC and DHC, Figure 4A) and the onset of enucleation (Figure 4C, black bars). The absence of dynein in late stages of erythroid maturation is consistent with proteomic studies of murine (Pasini et al., 2008) and human (Goodman et al., 2007) erythrocytes. In contrast, Trim58-deficient cells retained endogenous dynein protein aberrantly (Figures 4B and S5) and enucleation was delayed (Figure 4C, gray bars). Dynein protein levels still declined in Trim58 knockdown erythroblasts, but with an approximate 12 hour delay. The eventual degradation of most dynein by 44 hours in knockdown cells may be due to incomplete suppression of Trim58 or, more likely, alternate protein degradation mechanisms. Thus, expression of Trim58 destabilizes dynein and promotes enucleation, supporting a model in which these two processes are mechanistically linked.
Figure 4. Trim58 expression correlates with loss of dynein and enucleation during erythroid maturation.
Fetal liver erythroblasts were infected with retrovirus encoding shRNA against (A) luciferase (shLuc) or (B) Trim58 (shRNA #4), cultured in expansion medium for 72 hours, then shifted to maturation medium at time 0 (see also Figure 1D). Whole cell lysates were prepared at the indicated time points and endogenous proteins were analyzed by Western blotting. The lane marked “+” in panel B indicates endogenous Trim58 expression in shLuc-expressing cells at 44 hours. (C) Kinetics of enucleation in cells from panels A and B, determined as shown in Figure 1F. The results represent mean ± SD for 4 biological replicates. *p<0.05; **p<0.01; ns, not significant. See also Figure S5.
Trim58 regulates nuclear polarization
We used the ImageStream, which combines flow cytometry with fluorescent microscopy, to compare specific steps of enucleation in Trim58-depleted and control erythroblasts undergoing semi-synchronous maturation (Figure 5) (Konstantinidis et al., 2012). Erythroblasts infected with shRNA-containing retroviruses were identified by GFP expression and nuclei were stained with the cell-permeable DNA dye Draq5. We first measured changes in nuclear diameter over time to assess nuclear condensation during maturation (Figures 5A–D). No significant differences were observed between control and Trim58-depleted cells (Figure 5D), in agreement with independent morphometric analysis (Figure S2F). We next measured the aspect ratio (minor/major axis) and Δ centroid (distance between centers of the nucleus and cytoplasm) to distinguish spherical nucleated cells from oblong ones that were extruding their nuclei (Figures 5A–F). Spherical cells accumulated in Trim58-depleted cultures (Figure 5E), while the emergence of oblong cells undergoing nuclear extrusion was delayed (Figure 5F). Thus, Trim58 facilitates enucleation downstream of the nuclear condensation phase.
Figure 5. Trim58 regulates nuclear polarization and extrusion during erythropoiesis.
(A) Morphology parameters to assess specific steps of erythroid maturation by imaging flow cytometry. Declining nuclear diameter reflects condensation (broken line). The Δ centroid (distance between centers of the nucleus (red) and cytoplasm (green)) increases during nuclear polarization and extrusion. The aspect ratio (minor axis (blue line) divided by major axis (red line)) falls as cells become oblong during nuclear extrusion. (B) Representative analysis of fetal liver erythroblasts at 36 hours maturation. Spherical nucleated erythroblasts are pink. Oblong cells extruding their nuclei (decreased aspect ratio and increased Δ centroid) are orange. Cells in late mitosis, visualized to the left of the orange gate, exhibit low aspect ratio and low Δ centroid. (C) Representative images of cells from panel B that are, 1) spherical with a non-condensed centralized nucleus; 2) spherical with a condensed centralized nucleus; 3) spherical with a condensed polarized nucleus; or 4) oblong with a condensed extruding nucleus. (D–F) Erythroblasts expressing GFP and control (shLuc) or Trim58 shRNA #4 were analyzed by imaging flow cytometry at the indicated times during maturation. The results represent mean ± SD for 4 biological replicates, ~3000 cells analyzed per replicate. (D) Average nuclear diameter over time showing progressive condensation. (E) Percent (%) spherical cells over time, including those with central or polarized nuclei. (F) Percent (%) oblong cells with extruding nuclei over time. (G) Δ centroid distribution within spherical erythroblasts depicted in panel B. Higher Δ centroid values indicate polarized nuclei. Representative histograms are shown for three time points. Dashed white bars indicate the mean Δ centroid value for each plot. (H) Quantification of mean Δ centroid values in control (shLuc) or Trim58-deficient (shTrim58 #4) erythroblast cultures shown as mean ± SD for 3 biological replicates, ~3000 cells analyzed per replicate. *p<0.05, **p<0.01. See also Figure S6.
We assessed whether Trim58 regulated nuclear positioning, specifically within a population of spherical cells depicted in Figure 5E, by measuring their Δ centroid distribution (Figures 5G and 5H). At 32 and 36 hours maturation, the mean Δ centroid was significantly reduced in Trim58-depleted spherical cells, indicating impaired nuclear polarization. Treatment of control erythroblasts with the microtubule depolarizing agent nocodazole produced similar, albeit stronger effects (Figure S6), consistent with previous studies (Konstantinidis et al., 2012; Wang et al., 2012). Thus, erythroblast nuclear polarization is Trim58- and microtubule-dependent.
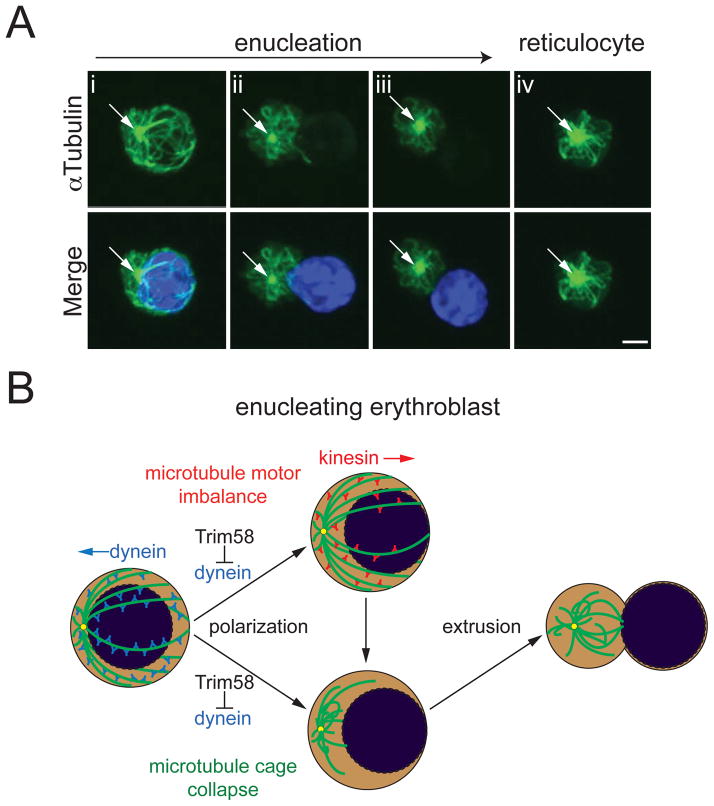
We used immunofluorescence to examine relationships between microtubule structure and nuclear positioning during enucleation. Erythroblast nuclei are surrounded by a network or “cage” of microtubules, similar to what occurs in most cells (Figure 6Ai) (Tsai et al., 2010; Wilson and Holzbaur, 2012). During enucleation, the nucleus moves away from a single microtubule-organizing center (MTOC) (Figure 6A and (Konstantinidis et al., 2012; Wang et al., 2012)), opposite to what would occur with minus end-directed dynein transport. Later, the microtubule cage partially collapses, becoming detached from the cell cortex and nucleus as the latter is extruded (Figures 6Aii-iv). Dynein stabilizes microtubules and tethers them to the cell cortex (Hendricks et al., 2012). Thus, events observed during enucleation, including directional nuclear movement and microtubule cage collapse (Figure 6B), may be facilitated by dynein loss (see also Figure 4A).
Figure 6. Nuclear movement during erythroblast enucleation.
(A) Primary murine fetal liver erythroblasts were fixed, stained for microtubules (αTubulin, green) and DNA (DAPI, blue), and analyzed by deconvolution fluorescent microscopy. Arrow indicates a single microtubule organizing center (MTOC). Scale bar, 2 μm. (B) Model for the actions of Trim58 during erythroblast enucleation. At early stages of maturation, the nucleus resides within a cage of microtubules (green), and is maintained in close proximity to the MTOC (yellow dot) by dynein. Trim58 causes dynein degradation, which promotes nuclear polarization through two potential mechanisms. First, microtubule motor imbalance may allow unopposed kinesin molecular motors to polarize the nucleus (top). Second, loss of dynein promotes detachment of microtubules from the nucleus and/or cell cortex (“cage collapse”), thereby enhancing polarization, extrusion and enucleation.
DISCUSSION
Genome wide association studies suggest that the E3 ubiquitin ligase superfamily member TRIM58 regulates human erythrocyte traits, including cell size and number (Kamatani et al., 2010; van der Harst et al., 2012). The current study supports a role for Trim58 in erythropoiesis and provides insight into associated mechanisms. Trim58 binds DIC directly, polyubiquitinates it in vitro, and induces proteasomal degradation of the dynein holocomplex in vivo. During erythropoiesis, Trim58 expression correlates with dynein loss and enucleation, both of which are inhibited by Trim58 depletion. Thus, we propose that Trim58 mediates erythroblast enucleation by interfering with the established ability of dynein to regulate nuclear positioning and/or microtubule structure within cells (Figure 6B) (McKenney et al., 2010; Splinter et al., 2010; Tanenbaum et al., 2011; Tsai et al., 2010; Wilson and Holzbaur, 2012). Our findings provide functional correlates for GWAS findings and identify a lineage-specific protein that alters the ubiquitous features of microtubule motor dynamics to regulate the specialized process of erythroblast enucleation.
According to our model (Figure 6B), delayed enucleation caused by Trim58 deficiency should be alleviated by inhibiting dynein through alternate mechanisms. However, our attempts to inhibit dynein in wild type and Trim58-deficient erythroblasts using dynein subunit shRNAs, overexpressed CC1 (Quintyne et al., 1999) or the chemical inhibitor ciliobrevin (Firestone et al., 2012) killed maturing erythroblasts prior to enucleation (data not shown). Similarly, retroviral expression of ectopic Trim58 in erythroblasts prior to their enucleation was toxic. These findings indicate that the timing and magnitude of endogenous Trim58 expression must be regulated precisely to occur exclusively during late erythropoiesis in order to preserve dynein-dependent processes required during earlier stages.
In most cells, networks or “cages” of microtubules surround the nucleus and regulate its spatial orientation through the actions of directionally opposed dynein and kinesin motors (Wilson and Holzbaur, 2012). During erythroblast enucleation, the nucleus moves away from the MTOC (Konstantinidis et al., 2012; Wang et al., 2012) and is ultimately released from the encompassing microtubule cage (Figure 6A). By degrading dynein, Trim58 could facilitate these processes via two potential mechanisms (Figure 6B). On one hand, movement of the nucleus during enucleation is directionally opposed to the minus end-directed motor actions of dynein. Thus, Trim58-mediated suppression of dynein may create a “microtubule motor imbalance”, allowing the plus end-directed actions of kinesins to polarize the nucleus away from the MTOC. Alternatively, dynein normally stabilizes microtubules and mediates their attachment to the cell cortex ((Hendricks et al., 2012) and references therein). Thus, Trim58-mediated dynein degradation in late stage erythroblasts may destabilize microtubules, promoting their detachment from the cell cortex and nuclear membrane (i.e., “microtubule cage collapse”) thereby facilitating nuclear movement and release. Of note, Trim58-directed shRNAs delayed, but did not prevent, dynein loss and enucleation during late erythropoiesis (see Figure 4). This may be explained by incomplete suppression of Trim58. Furthermore, redundant systems, including other erythroid ubiquitin ligases, autophagy, and/or proteases, may cooperate with Trim58 to degrade dynein. This possibility is consistent with the extensive repertoire of proteolytic systems present in late stage erythroblasts (Khandros and Weiss, 2010).
Kinesin-1 regulates nuclear movement in numerous cell types (Tanenbaum et al., 2011) and its Kif5b isoform is abundant in murine (Kingsley et al., 2013) and human (Merryweather-Clarke et al., 2011) erythroblasts. Knockdown of Kif5b by shRNAs did not antagonize the deleterious effects of Trim58 deficiency on enucleation (data not shown), as would be predicted by the “microtubule motor imbalance” model. This finding supports the alternative mechanism of “microtubule cage collapse”. However, at least 10 additional kinesin family members are expressed during erythropoiesis ((Kingsley et al., 2013; Merryweather-Clarke et al., 2011) and data not shown). Systematic inhibition of these kinesins, either separately or in combinations, may determine whether any participate in nuclear polarization during erythropoiesis.
Several lines of evidence suggest important roles for ubiquitin proteasome-mediated proteolysis during erythroblast maturation (Khandros and Weiss, 2010) and enucleation (Chen et al., 2002), but only a few specific pathways are defined. For example, the generally expressed ubiquitin ligases Mdm2 and Mdm4 foster erythropoiesis by inhibiting p53 (Maetens et al., 2007), and the ubiquitin ligase Cul4A regulates erythropoiesis by targeting the cell cycle inhibitor p27 (Li et al., 2006). Trim10/Herf1 (Harada et al., 1999), E2-20K and E2-230K (Wefes et al., 1995) are erythroid-enriched ubiquitin ligases of unknown function. Here, we show that Trim58 is a functionally important ubiquitin ligase in erythroid cells and identify dynein as a biologically relevant substrate. In principle, Trim58 could also interfere with dynein activity by displacing other proteins that interact with the DIC amino-terminal coiled coil domain, including dynactin/p150Glued and NudE (McKenney et al., 2011). However, overexpression of the RΔ Trim58 mutant, which lacks ubiquitin ligase activity but still interacts with DIC, did not elicit phenotypes characteristic of dynein loss in HeLa cells (Figures 3E–G).
It is also possible that Trim58 promotes erythroid maturation via dynein-independent mechanisms. For example, IP studies demonstrated Trim58 interactions with Nup50, Nup153, importin α and importin β (Table S1), all of which can coexist in a complex (Makise et al., 2012; Matsuura and Stewart, 2005). Trim58 may interact with this complex in post-mitotic erythroblasts, for example, through the C-terminus of Nup153, which has been localized to the cytoplasmic face of the nuclear pore complex (Fahrenkrog et al., 2002). Moreover, some nuclear pore proteins physically redistribute during erythropoiesis, which could enhance their accessibility to Trim58 (Krauss et al., 2005), Trim58 may also promote erythroid-specific restructuring of the nuclear pore, either by acting as a scaffold or via ubiquitination-mediated effects on protein localization. Such effects may be proteasome-independent, as Trim58 does not appear to facilitate downregulation of Nup153 (Figures 4A and 4B and data not shown).
Genome wide association studies offer a population-based approach to identify genes that regulate health-related traits. However, GWAS typically identify single nucleotide polymorphisms within linkage disequilibrium blocks that contain multiple genes, and it can be challenging to identify the specific gene(s) that influences the trait of interest (Hindorff et al., 2009; Peters and Musunuru, 2012). Moreover, alleles discovered by GWAS usually cause subtle alterations in gene expression or function, providing a minimal estimate of the role for a particular gene in the biological process of interest (Sankaran and Orkin, 2013). Our Trim58 overexpression and loss-of-function studies address these general issues by demonstrating a specific role for Trim58 in erythroid cells and by identifying a relevant molecular pathway. GWAS also identify TRIM58 as a candidate gene for regulating human platelet numbers (Gieger et al., 2011). Trim58 mRNA is induced in megakaryocytes, likely via GATA1 and SCL/Tal1 (Figures S1F and S1G), similar to what we observed in erythroid cells. These observations are interesting in light of findings that dynein promotes platelet production by regulating microtubule dynamics in megakaryocytes (Patel et al., 2005). Thus, Trim58 may regulate platelet formation through its interactions with dynein. Overall, our findings illustrate how mechanism-based investigations synergize with GWAS to elucidate new biological pathways.
Considerable efforts have been directed toward understanding how dynein activities are regulated in different cellular contexts. Dynein cargo preference and motor activity are modulated largely by protein interactions and perhaps by post-translational modifications (Vallee et al., 2012). Our findings demonstrate that dynein is also regulated by precisely timed, lineage-specific degradation. It will be interesting to explore the generality of this finding by investigating whether dynein stability is regulated in non-erythroid tissues through interactions with Trim58 or other functionally related ubiquitin ligases.
EXPERIMENTAL PROCEDURES
Trim58 cloning
Trim58 cDNA was initially isolated from a primary murine fetal liver erythroblast cDNA library and cloned into various expression vectors. See Extended Experimental Procedures for primer sequences.
Murine fetal liver erythroblast assay
Erythroid precursors were purified cultured and infected with shRNA-expressing retrovirus as described (Khandros et al., 2012). For flow cytometry, 5 × 105 cultured fetal liver erythroblasts were stained sequentially with 5 μM Hoescht 33342 (Sigma) for 1 hour at 37 °C in fetal liver maturation medium, LiveDead near-IR fixable dead cell stain (Invitrogen) for 30 min at 4 °C in phosphate buffered saline, pH 7.5 (PBS), and Ter119-PerCP-Cy5.5 and CD44-AF647 (BioLegend) for 45 min at 4 °C in PBS with 2% fetal bovine serum (FBS). Cells were analyzed on an LSR Fortessa flow cytometer (BD Biosciences).
Flow cytometry data were analyzed using FlowJo software (TreeStar). Cell debris and free pyrenocytes (Forward Scatterlow, Hoechst+) were excluded from the total cell population prior to analysis. Cell viability was calculated as the percent live cells divided by the total number of cells. Enucleation was calculated as the percent live, anucleate reticulocytes (Hoechst−/Forward Scatterlow) divided by the total number of live cells. See Extended Experimental Procedures for further protocol details and short hairpin RNA sequences.
Anti-Trim58 antibody generation
Anti-murine Trim58 antibodies were raised against a N-terminal peptide representing amino acids 7–29 (ERLQEEARCSVCLDFLQEPISVD) (Thermo Scientific).
Mass spectrometry
Immunoprecipitates were size-fractionated by SDS-PAGE (4–15% gradient gel, BioRad), stained with Coomassie Blue Silver overnight and destained in deionized water. The prominent visible protein bands were manually extracted, digested with trypsin, and subjected to liquid chromatography and nanospray/linear trap quadrupole mass spectrometry using a ThermoFinnigan LTQ linear ion trap mass spectrometer at the University of Pennsylvania Proteomics Core Facility. Data were analyzed using Sequest and Scaffold3 software packages. The data shown in Tables 1 and S1 include proteins identified by ≥2 peptides with >99% protein/>95% peptide confidence.
SEC-MALLS experiments
Recombinant proteins were produced separately from E. coli Rosetta 2 (DE3) cells and run individually or as a mixture on a Superdex 200 10/300 GL column connected to a MiniDawn Treos (Wyatt Technology). See Extended Experimental Procedures for purification techniques and further experimental details.
In vitro ubiquitination assays
Recombinant E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme (UBE2D3), and Trim63 were purchased from Boston Biochem. Full length Trim58 proteins were produced as GST fusions in E. coli BL21 and purified from inclusion bodies using Sarkosyl extraction (Tao et al., 2010). Ubiquitination reactions (20 μL) contained Mg-ATP (2 mM), 300 nM GST-Trim58 or GST-Trim63 protein as positive control, HA-tagged ubiquitin (1 μg), E1 (50 nM), and E2 (1 μM). In some experiments, 100 μM recombinant purified GST-DIC (1–120) was added. Reactions were incubated for 1 hour at 37 °C in ubiquitination buffer (50 mM HEPES pH 8, 50 mM sodium chloride, and 1 mM Tris 2-carboxyethyl phosphine) and terminated by addition of an equal volume of 2X Laemmli buffer and heating at 95 °C for 5 min. The reactions were resolved by SDS-PAGE (4–20% polyacrylamide) and analyzed by Western blotting using monoclonal HA, monoclonal GST, or polyclonal DIC antibodies. Blots were incubated in denaturing buffer (6 M guanidine HCl, 20 mM Tris-HCl pH 7.4, 1 mM PMSF, and 5 mM β-mercaptoethanol) for 30 min at 4 °C before the blocking step.
Immunofluorescence and time lapse microscopy
HeLa cells and erythroblasts were prepared for imaging using standard techniques (see Extended Experimental Procedures). Imaging was conducted on microscopes maintained by the University of Pennsylvania Perelman School of Medicine Cell and Developmental Biology Microscopy Core.
Amnis ImageStream flow cytometry
Erythroid cultures were fixed in 4% paraformaldehyde for 15 min at room temperature, washed in PBS, stained with Draq5 (Abcam), and analyzed on an Amnis ImageStream Mark II instrument at 40x magnification. GFP+ cells were gated using IDEAS software (Amnis) and comprised the base population for morphologic analysis based on aspect ratio and Δ centroid values. “Spherical” and “Oblong” cell gates were set by manual inspection of fluorescent cell images. We designed a MATLAB program to identify nucleated cells within the spherical cell population by excluding anucleate reticulocytes (Draq5-negative) and multinucleated cells (detected by multiple Draq5-positive regions). Nuclear diameter, measured by Draq5 staining, was quantified in MATLAB. IDEAS software was used to calculate Δ centroid values within the Draq5-positive, spherical cell population for assessing nuclear polarization. For some experiments, 16 μM nocodazole (Sigma) was added directly to culture media for the indicated times.
Statistics
Statistical analysis was performed using GraphPad Prism 6.0 software (GraphPad Software). All multigroup comparisons were done using one-way ANOVA analysis. Comparisons between two groups were done using Student’s t-test. Significance was set at p<0.05.
Supplementary Material
HIGHLIGHTS.
Genome wide association studies link the TRIM58 gene to human erythrocyte traits.
Trim58 shRNAs delay normal enucleation of mouse erythroblasts.
Trim58 binds the molecular motor dynein and promotes its proteasomal degradation.
Trim58-mediated elimination of dynein may promote erythroblast enucleation.
Acknowledgments
We thank Gerd Blobel, Laura Gutierrez, Mark Kahn, Katherine Nathanson, Stephan Kadauke, Katherine Ullman, Katherine High and Xiaolu Yang for helpful conversations and reagents. We thank Mariko Tokito and Patricia Mericko-Ishizuka for technical assistance. Min Min Lu at the University of Pennsylvania Molecular Cardiology Research Center Histology and Gene Expression Core provided assistance with in situ hybridization experiments. Andrea Stout at the University of Pennsylvania Cell and Developmental Biology Microscopy Core provided assistance with microscopy. This work was funded by NIH grants P30DK090969 (MJW), DK61692 (MJW), GM007170 (CST, EAT), HL007439 (CST, EAT), and GM48661 (ELFH).
Footnotes
AUTHOR CONTRIBUTIONS
CST, EAT, EK and MJW conceived of the project. CST, EAT, EK, JMN, OYZ, DP, YY, CA and MJW designed and conducted experiments, and analyzed data. JEL and ELFH provided purified holodynein and assisted in conducting and analyzing experiments. APGS and JPM performed the SEC-MALLS studies. SYF helped to conduct and analyze in vitro ubiquitination experiments. CST, EAT and MJW wrote the paper.
Supplemental Information includes Extended Experimental Procedures, six figures, four movies, and one table.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen CY, Pajak L, Tamburlin J, Bofinger D, Koury ST. The effect of proteasome inhibitors on mammalian erythroid terminal differentiation. Exp Hematol. 2002;30:634–639. doi: 10.1016/s0301-472x(02)00826-3. [DOI] [PubMed] [Google Scholar]
- Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolznig H, Bartunek P, Nasmyth K, Mullner EW, Beug H. Terminal differentiation of normal chicken erythroid progenitors: shortening of G1 correlates with loss of D-cyclin/cdk4 expression and altered cell size control. Cell Growth Differ. 1995;6:1341–1352. [PubMed] [Google Scholar]
- Fabrini R, De Luca A, Stella L, Mei G, Orioni B, Ciccone S, Federici G, Lo Bello M, Ricci G. Monomer-dimer equilibrium in glutathione transferases: a critical re-examination. Biochemistry. 2009;48:10473–10482. doi: 10.1021/bi901238t. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O’Donnell M, Gelfand VI, Kapoor TM, Chen JK. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaehtgens P, Schmidt F, Will G. Comparative rheology of nucleated and non-nucleated red blood cells. I. Microrheology of avian erythrocytes during capillary flow. Pflugers Arch. 1981;390:278–282. doi: 10.1007/BF00658276. [DOI] [PubMed] [Google Scholar]
- Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, Serbanovic-Canic J, Elling U, Goodall AH, Labrune Y, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SC, Derganc J, Shevkoplyas SS, Yoshida T, Bitensky MW. A detailed study of time-dependent changes in human red blood cells: from reticulocyte maturation to erythrocyte senescence. Br J Haematol. 2006;135:395–404. doi: 10.1111/j.1365-2141.2006.06279.x. [DOI] [PubMed] [Google Scholar]
- Goodman SR, Kurdia A, Ammann L, Kakhniashvili D, Daescu O. The human red blood cell proteome and interactome. Exp Biol Med (Maywood) 2007;232:1391–1408. doi: 10.3181/0706-MR-156. [DOI] [PubMed] [Google Scholar]
- Harada H, Harada Y, O’Brien DP, Rice DS, Naeve CW, Downing JR. HERF1, a novel hematopoiesis-specific RING finger protein, is required for terminal differentiation of erythroid cells. Mol Cell Biol. 1999;19:3808–3815. doi: 10.1128/mcb.19.5.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur EL. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Clifford M, Losson R, Tanabe O, Engel JD. TRIM28 is essential for erythroblast differentiation in the mouse. Blood. 2013;122:3798–3807. doi: 10.1182/blood-2013-04-496166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci U S A. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayapal SR, Lee KL, Ji P, Kaldis P, Lim B, Lodish HF. Down-regulation of Myc is essential for terminal erythroid maturation. J Biol Chem. 2010;285:40252–40265. doi: 10.1074/jbc.M110.181073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol. 2008;10:314–321. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- Ji P, Yeh V, Ramirez T, Murata-Hori M, Lodish HF. Histone deacetylase 2 is required for chromatin condensation and subsequent enucleation of cultured mouse fetal erythroblasts. Haematologica. 2010;95:2013–2021. doi: 10.3324/haematol.2010.029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- Keerthivasan G, Small S, Liu H, Wickrema A, Crispino JD. Vesicle trafficking plays a novel role in erythroblast enucleation. Blood. 2010;116:3331–3340. doi: 10.1182/blood-2010-03-277426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthivasan G, Wickrema A, Crispino JD. Erythroblast enucleation. Stem Cells Int. 2011;2011:139851. doi: 10.4061/2011/139851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandros E, Thom CS, D’Souza J, Weiss MJ. Integrated protein quality-control pathways regulate free alpha-globin in murine beta-thalassemia. Blood. 2012;119:5265–5275. doi: 10.1182/blood-2011-12-397729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandros E, Weiss MJ. Protein quality control during erythropoiesis and hemoglobin synthesis. Hematol Oncol Clin North Am. 2010;24:1071–1088. doi: 10.1016/j.hoc.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley PD, Greenfest-Allen E, Frame JM, Bushnell TP, Malik J, McGrath KE, Stoeckert CJ, Palis J. Ontogeny of erythroid gene expression. Blood. 2013;121:e5–e13. doi: 10.1182/blood-2012-04-422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis DG, Pushkaran S, Johnson JF, Cancelas JA, Manganaris S, Harris CE, Williams DA, Zheng Y, Kalfa TA. Signaling and cytoskeletal requirements in erythroblast enucleation. Blood. 2012;119:6118–6127. doi: 10.1182/blood-2011-09-379263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss SW, Lo AJ, Short SA, Koury MJ, Mohandas N, Chasis JA. Nuclear substructure reorganization during late-stage erythropoiesis is selective and does not involve caspase cleavage of major nuclear substructural proteins. Blood. 2005;106:2200–2205. doi: 10.1182/blood-2005-04-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Jia N, Kapur R, Chun KT. Cul4A targets p27 for degradation and regulates proliferation, cell cycle exit, and differentiation during erythropoiesis. Blood. 2006;107:4291–4299. doi: 10.1182/blood-2005-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Guo X, Mohandas N, Chasis JA, An X. Membrane remodeling during reticulocyte maturation. Blood. 2010;115:2021–2027. doi: 10.1182/blood-2009-08-241182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Triviños-Lagos L, Gräf R, Chisholm RL. Dynein Intermediate Chain Mediated Dynein–Dynactin Interaction Is Required for Interphase Microtubule Organization and Centrosome Replication and Separation in Dictyostelium. J Cell Biol. 1999;147:1261–1274. doi: 10.1083/jcb.147.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, Klingmuller U, Lozano G, Marine JC. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. The Nup153-Nup50 interface and its role in nuclear import. J Cell Biol. 2012;289:38515–38522. doi: 10.1074/jbc.M112.378893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr Top Dev Biol. 2008;82:1–22. doi: 10.1016/S0070-2153(07)00001-4. [DOI] [PubMed] [Google Scholar]
- McGrath KE, Kingsley PD, Koniski AD, Porter RL, Bushnell TP, Palis J. Enucleation of primitive erythroid cells generates a transient population of “pyrenocytes” in the mammalian fetus. Blood. 2008;111:2409–2417. doi: 10.1182/blood-2007-08-107581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryweather-Clarke AT, Atzberger A, Soneji S, Gray N, Clark K, Waugh C, McGowan SJ, Taylor S, Nandi AK, Wood WG, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011;117:e96–108. doi: 10.1182/blood-2010-07-290825. [DOI] [PubMed] [Google Scholar]
- Mueller RL, Gregory TR, Gregory SM, Hsieh A, Boore JL. Genome size, cell size, and the evolution of enucleated erythrocytes in attenuate salamanders. Zoology (Jena) 2008;111:218–230. doi: 10.1016/j.zool.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano LM, Meroni G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- Pasini EM, Kirkegaard M, Salerno D, Mortensen P, Mann M, Thomas AW. Deep coverage mouse red blood cell proteome: a first comparison with the human red blood cell. Mol Cell Proteomics. 2008;7:1317–1330. doi: 10.1074/mcp.M700458-MCP200. [DOI] [PubMed] [Google Scholar]
- Patel SR, Richardson JL, Schulze H, Kahle E, Galjart N, Drabek K, Shivdasani RA, Hartwig JH, Italiano JE., Jr Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood. 2005;106:4076–4085. doi: 10.1182/blood-2005-06-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DT, Musunuru K. Functional evaluation of genetic variation in complex human traits. Hum Mol Genet. 2012;21:R18–23. doi: 10.1093/hmg/dds363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M, Kossenkov AV, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC, et al. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res. 2014 doi: 10.1101/gr.164178.113. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop R, Shearstone JR, Shen Q, Liu Y, Hallstrom K, Koulnis M, Gribnau J, Socolovsky M. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010;8:e1000484. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Krauss SW, Short SA, Lee G, Villalobos J, Etzell J, Koury MJ, Ney PA, Chasis JA, Grigoryev SA. Chromatin condensation in terminally differentiating mouse erythroblasts does not involve special architectural proteins but depends on histone deacetylation. Chromosome Res. 2009;17:47–64. doi: 10.1007/s10577-008-9005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Orkin SH. Genome-wide association studies of hematologic phenotypes: a window into human hematopoiesis. Curr Opin Genet Dev. 2013;23:339–344. doi: 10.1016/j.gde.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni S, Bala S, Gwynn B, Sahr KE, Peters LL, Hanspal M. Absence of erythroblast macrophage protein (Emp) leads to failure of erythroblast nuclear extrusion. J Biol Chem. 2006;281:20181–20189. doi: 10.1074/jbc.M603226200. [DOI] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Akhmanova A, Medema RH. Bi-directional transport of the nucleus by dynein and kinesin-1. Commun Integr Biol. 2011;4:21–25. doi: 10.4161/cib.4.1.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Liu W, Simmons BN, Harris HK, Cox TC, Massiah MA. Purifying natively folded proteins from inclusion bodies using sarkosyl, Triton X-100, and CHAPS. Biotechniques. 2010;48:61–64. doi: 10.2144/000113304. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, Paul DS, Elling U, Allayee H, Li X, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ramirez T, Ji P, Jayapal SR, Lodish HF, Murata-Hori M. Mammalian erythroblast enucleation requires PI3K-dependent cell polarization. J Cell Sci. 2012;125:340–349. doi: 10.1242/jcs.088286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefes I, Mastrandrea LD, Haldeman M, Koury ST, Tamburlin J, Pickart CM, Finley D. Induction of ubiquitin-conjugating enzymes during terminal erythroid differentiation. Proc Natl Acad Sci U S A. 1995;92:4982–4986. doi: 10.1073/pnas.92.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Holzbaur EL. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci. 2012;125:4158–4169. doi: 10.1242/jcs.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006;24:967–976. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nat Cell Biol. 2005;437:754–758. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang NJ, Chen C, Tang W, Kornbluth S. Ubiquitylation of p53 by the APC/C inhibitor Trim39. Proc Natl Acad Sci U S A. 2012;109:20931–20936. doi: 10.1073/pnas.1212047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.