Fig. 3. Collaborative model of epigenomic lineage determination in macrophages.
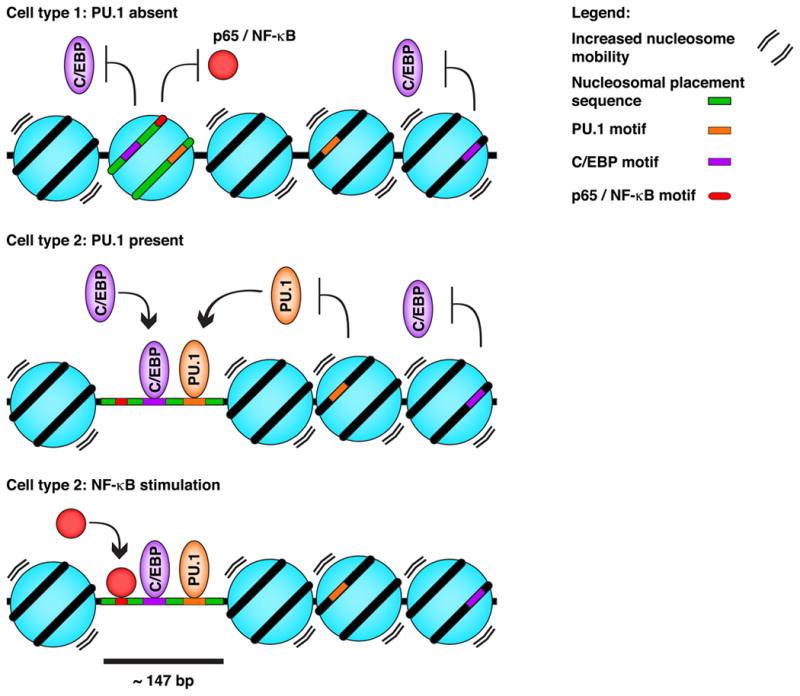
Co-occurrences of binding sequences for PU.1 and collaborative TFs, including C/EBP factors, within a ∼147 nucleotides window are relatively prevalently encoded within larger DNA sequences that promote a more defined nucleosome placement (in green). In cells that do not express PU.1, such specific nucleosome placement may protect these cells from using genomic information not relevant to their functions. Note that neighboring nucleosomes may be more mobile relative to their interaction with DNA (arrowheads). In macrophages however, the same nucleosome is probabilistically more effectively evicted and/or displaced as a result of the collaborative binding activity of PU.1 and C/EBP factors. This then allows for more efficient binding of the NF-κB p65 subunit following TLR4 stimulation, for example.
