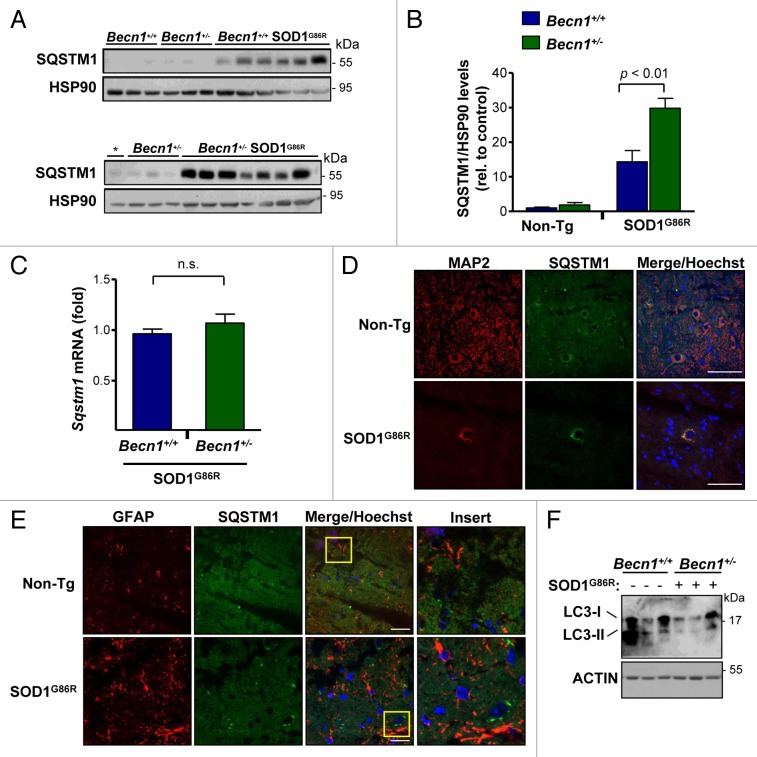
Figure 4. Altered levels of autophagy markers in transgenic SOD1 mice heterozygous for Becn1. (A) Levels of SQSTM1 were monitored in spinal cord extracts of symptomatic SOD1G86R transgenic mice by western blot and quantified by densitometry. Each lane represents a single mouse, ordered by genotype and then by life span (incremented values). “*” represents the same sample from the first lane of a Becn1+/+ SOD1G86R mouse shown in the left gel used as a normalization sample. (B) SQSTM1 levels were quantified from samples described in (A). Statistical analysis was performed using a one-way ANOVA test; data represent mean and standard error of at least 4 independent animals per genotype. (C) Sqstm1 mRNA levels were measured by real-time PCR in samples presented in (A). Mean and standard error is shown. n.s., nonsignificant differences. (D) Immunofluorescence analysis of SQSTM1 (green) and MAP2 staining (neuronal marker, red) or (E) SQSTM1 and GFAP staining (astrocytes, red) was performed in spinal cord tissue derived from SOD1G86R and non-transgenic littermate control mice (Non-Tg). Hoechst staining was also performed (blue). A merged image of the triple staining is presented together with a zoom of the selected area (yellow square). A representative image is presented of the analysis of 3 independent animals for genotype. Scale bars: 10 µm. (F) Levels of LC3-I and II were evaluated by western blot in spinal cord samples from Becn1+/+ SOD1G86R and Becn1+/− SOD1G86R mice. The western blot for LC3-I and II was performed in a 17% SDS-PAGE. Each well represents an independent animal analyzed.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.