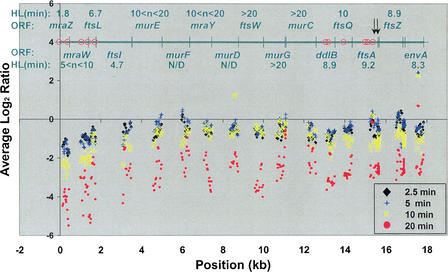
Figure 3.
High-resolution analysis of the dcw gene cluster transcripts. Average log2 ratios (y-axis) were plotted against operon position (x-axis) for three-probe sliding windows of the 156 probes in the dcw gene cluster, including 30 bases up- and downstream of the first and last ORFs. The positions of known promoters (red circles) and the ORFs with their estimated half-lives are given in the upper part of the graph. Arrows indicate known RNase E processing sites. An additional weak promoter is thought to be present in either murD or ftsW (Mengin-Lecreulx et al. 1998). rho-independent terminators are present in the 5′ region of mraZ and immediately downstream of envA. Degradation is fastest at the 5′ end of the operon, with three apparent regions of distinct degradation rates: The 5′ region (mraZ–ftsI) is degraded the fastest, the middle (murE–murC) is relatively stable, and the 3′ region (ddlB–envA) is degraded at an intermediate rate.