Abstract
Centromeres are the site for kinetochore formation and spindle attachment and are embedded in heterochromatin in most eukaryotes. The repeat-rich nature of heterochromatin has hindered obtaining a detailed understanding of the composition and organization of heterochromatic and centromeric DNA sequences. Here, we report the results of extensive sequence analysis of a fully functional centromere present in the Drosophila Dp1187 minichromosome. Approximately 8.4% (31 kb) of the highly repeated satellite DNA (AATAT and TTCTC) was sequenced, representing the largest data set of Drosophila satellite DNA sequence to date. Sequence analysis revealed that the orientation of the arrays is uniform and that individual repeats within the arrays mostly differ by rare, single-base polymorphisms. The entire complex DNA component of this centromere (69.7 kb) was sequenced and assembled. The 39-kb “complex island” Maupiti contains long stretches of a complex A+T rich repeat interspersed with transposon fragments, and most of these elements are organized as direct repeats. Surprisingly, five single, intact transposons are directly inserted at different locations in the AATAT satellite arrays. We find no evidence for centromere-specific sequences within this centromere, providing further evidence for sequence-independent, epigenetic determination of centromere identity and function in higher eukaryotes. Our results also demonstrate that the sequence composition and organization of large regions of centric heterochromatin can be determined, despite the presence of repeated DNA.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos.: Beagle = AY183918, F = AY183919, 412 = AY183920, Bel = AY183921, You = AY183922, Maupiti = AY183926, AATAT = AY183925, AY183931–AY184007, and TTCTC = AY183923–AY183924, AY183927–AY183930, AY184008–AY184069].
The last few years have witnessed the publication of complete or nearly complete euchromatic physical maps and sequence assemblies for Caenorhabditis elegans, Arabidopsis thaliana, Drosophila melanogaster, and most recently, Homo sapiens (The C. elegans Sequencing Consortium 1998; Adams et al. 2000; The Arabidopsis Genome Initiative 2000; Lander et al. 2001; Venter et al. 2001). The heterochromatin comprises ∼30% of both the fly and human genomes, yet it has been virtually ignored by these large-scale genome projects. This enigmatic part of the genome has unusual cytological, molecular, and genetic properties, including differential control of replication, condensation throughout the cell cycle, and the ability to silence gene expression (John 1988; Weiler and Wakimoto 1996; Elgin and Workman 2002). Heterochromatin is concentrated in large (megabase-sized) blocks, predominantly in the centric and subtelomeric regions of all chromosomes, and contains tandemly repeated short sequences (satellite DNAs), middle repetitive elements (e.g., transposons), and some single-copy DNA.
Heterochromatin has been unflatteringly referred to as “junk DNA,” because it is more difficult to assign functions to repeated sequences than to protein-encoding sequences. However, heterochromatin is not inert and has been demonstrated to be essential for cell and organismal viability in multicellular eukaryotes. Essential genes (e.g., lethal mutable genes, ribosomal RNA genes) and fertility genes (e.g., Y-linked male fertility factors) reside in heterochromatin (Gatti and Pimpinelli 1992). Essential cis-acting chromosome inheritance functions are also mediated by heterochromatin, including centromeres (Sullivan et al. 2001), meiotic pairing (McKee and Karpen 1990; Dernburg et al. 1996; Karpen et al. 1996), and sister chromatid cohesion (Moore and Orr-Weaver 1998; Bernard et al. 2001). Thus, a complete understanding of eukaryotic genomes requires detailed structural and functional analysis of heterochromatin.
One essential heterochromatic element is the centromere, which is associated with the kinetochore. The centromere/kinetochore complex is necessary for spindle attachment, prophase and anaphase chromosome movements, and the activity of the spindle assembly checkpoint (SAC) (for review, see Dobie et al. 1999; Sullivan et al. 2001). One key question currently under debate is what determines centromere identity. How is only one site (in most eukaryotes) chosen for centromere function and kinetochore formation? Cytological and biochemical studies have demonstrated a physical association between tandemly repeated satellite DNAs and centromere regions and proteins in higher eukaryotes (Willard 1990; Vafa and Sullivan 1997; Sullivan 2001; Sullivan et al. 2001). However, normal centromeric DNA is neither necessary nor sufficient for centromere function (Karpen and Allshire 1997; Choo 2000; Sullivan et al. 2001). Different heterochromatic properties and functions have been demonstrated to be regulated in a sequence-independent manner by epigenetic mechanisms (Allshire et al. 1994; Hendrich and Willard 1995; Wakimoto 1998; Jenuwein and Allis 2001). In comparison to models that emphasize the importance of primary DNA sequence to centromere identity and propagation, epigenetic models parsimoniously account for both the stability and plasticity of centromeres (Karpen and Allshire 1997; Choo 2000; Sullivan et al. 2001).
The involvement of epigenetic regulation does not preclude a role for DNA sequence in centromere identity or function, or other functions encoded by heterochromatin. For example, secondary sequence characteristics, such as A+T richness, or the ability to form higher-order structures through bending, may facilitate the conversion of an epigenetic mark into the formation of a functional kinetochore (Vig 1994; Murphy and Karpen 1998; Koch 2000; Sullivan et al. 2001). In addition, specific sequences may serve as boundary elements that constrain the spreading of centromeric chromatin (Maggert and Karpen 2001; Blower et al. 2002). Thus, understanding heterochromatic and centromeric sequences would reveal insights into their functional regulation, in addition to providing a more complete understanding of genome structure and sequence.
Repetitive DNA poses significant challenges to cloning, sequencing, and assembly. Nevertheless, substantial progress has been made in the analysis of the nonsatellite component of Drosophila, Arabidopsis, and human heterochromatin (Copenhaver et al. 1999; Kotani et al. 1999; Adams et al. 2000; The Arabidopsis Genome Initiative 2000; Horvath et al. 2000a,b; Kumekawa et al. 2000; Haupt et al. 2001). These regions predominantly contain transposons and transposon fragments, as well as genes and gene fragments. Less progress has been made in genomic analysis of satellite sequences. The nucleotide composition of individual repeats has been obtained from sequencing small clones of satellite DNA (Lohe and Brutlag 1986; Jabs and Persico 1987; Warburton et al. 1996; Losada et al. 1997; Haaf and Willard 1998). In situ hybridization to mitotic chromosomes, along with classical cytogenetic banding methods, have revealed the gross distribution of some repeated DNAs within heterochromatin in organisms such as Drosophila (Lohe et al. 1993; Pimpinelli et al. 1995) and humans (Dunham et al. 1992; Grady et al. 1992; Mullenbach et al. 1996). However, cytological methods do not provide high-resolution information about the size and complexity of simple sequence arrays. Although pulsed field gel electrophoresis (PFGE) provides one method for spanning the gap between short-satellite sequences and cytological mapping (Sun et al. 1997), the dispersion of repetitive DNAs throughout the genome, and tandem repetition of satellites, has impeded extensive restriction mapping of specific heterochromatic regions. Chromosome-specific satellite DNAs and PFGE have been used to successfully map satellite domains in mammals; however, these maps are limited by the lack of direct molecular access deep within individual satellite blocks (Willard et al. 1986; Jabs and Persico 1987; Jabs et al. 1989; Arn et al. 1991; Schueler et al. 2001).
Recent studies have revealed significant, unprecedented information about centromere region sequences. Detailed sequence analyses of human centromeres have been published recently, revealing the substructure and composition of the satellite arrays (Lee et al. 2000; Schueler et al. 2001). However, the sequence assemblies of these regions of satellite DNA are not complete, leaving open the possibility that other types of sequences are present in the arrays. In Arabidopsis, the pericentromeric regions on all five chromosomes have been cloned and sequenced (Copenhaver et al. 1999; Kotani et al. 1999; The Arabidopsis Genome Initiative 2000; Kumekawa et al. 2000; Haupt et al. 2001). Detailed studies of some of these pericentromeric regions demonstrate that they contain intact and fragmented transposons as well as single-copy genes. However, the satellite regions present in Arabidopsis centromere regions have not been completely sequenced. Although we have a better understanding of the organization and composition of some higher eukaryotic centromere regions, we lack an understanding of the exact primary sequence of any functional, higher-eukaryotic centromere. Similarly, a number of important questions about the global and detailed organization of heterochromatic DNA, and the relationship between heterochromatin sequences and functions, remain unanswered.
We have studied the molecular genetics of chromosome structure and inheritance using the Drosophila minichromosome Dp1187. This minichromosome contains the components and inheritance functions associated with normal higher eukaryotic chromosomes, yet is amenable to molecular analysis and manipulation. It is relatively small (1.3 Mb, or ∼1/30th the size of the normal X chromosome), contains easily scorable genetic markers, and is not essential for the viability of the cell or organism. Studies utilizing this minichromosome system have generated information about chromosome structure, and the cis and trans regulators of chromosome inheritance, nuclear organization, and gene expression (e.g., Karpen and Spradling 1990; Murphy and Karpen 1995a; Karpen et al. 1996; Dobie et al. 2001; Hari et al. 2001; Maggert and Karpen 2001; Donaldson et al. 2002). Analysis of the transmission behavior of molecularly defined Dp1187 deletion derivatives localized the sequences necessary for chromosome inheritance to within a specific 420-kb portion of the centric heterochromatin (Murphy and Karpen 1995b). Pulsed-field Southern analysis revealed the gross composition and organization of this functional centromere, as well as the rest of the minichromosome centric heterochromatin (Sun et al. 1997). These studies revealed the presence of two large blocks of simple, highly repeated satellite sequences in the functional centromere, as well as complex DNA in the form of intact and fragmented transposons.
We wanted to gain a more precise understanding of the sequence composition and organization of centric heterochromatin and specifically of a centromere demonstrated to function in vivo. Therefore, we have cloned, sequenced, and assembled all the complex DNA within the Dp1187 centromere, and 8.4% of the satellite arrays. Here, we describe the results of these studies, which demonstrate that centric heterochromatin sequencing and assembly can be accomplished. Furthermore, these studies confirm the uniformity of the satellite arrays, reveal that the transposons have very high identity to euchromatic copies, and show that the structure of one end of the centromere is complex, and includes higher-order repeat structures. We discuss the relevance of these findings to the evolution of heterochromatin and to current models for the determination of centromere identity, and discuss the possibility that genome projects can ultimately be truly completed through inclusion of assembled heterochromatic sequences.
RESULTS
Generation of a Centromere-Enriched Library From Gel-Purified Minichromosome DNA
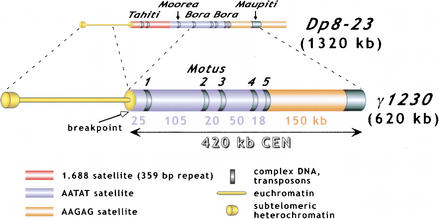
The presence of the same repeats in different parts of the genome is the major impediment to mapping and sequencing a specific region of heterochromatin. To circumvent these problems, we have used gel purification of a minichromosome derivative, allowing us to focus cloning and sequencing efforts on one specific region of centric heterochromatin, namely a genetically defined functional centromere. The only centric heterochromatin in the 620-kb Dp1187 derivative γ1230 corresponds to the 420-kb, fully functional centromere (Fig. 1). Previously, PFGE of gel-purified γ1230 DNA was used to restriction map the centromere, which revealed its gross organization and composition (Fig. 1)(Sun et al. 1997). This analysis identified and positioned two large blocks of AATAT and AAGAG satellites, five islands of complex DNA (Motus 1–5, Tahitian for “small island”) embedded in the AATAT array, and a large complex island at the right end of the centromere (Maupiti). Hybridization analysis and restriction mapping suggested that Motus 1–4 contained known transposable elements: three Long Terminal Repeat (LTR)-containing retrotransposons (HMS Beagle, 412, and Belshazar/3S18), plus one non-LTR (LINE-like) retroposon (F).
Figure 1.
Origin and structure of γ1230. γ1230 was generated by radiation-induced deletions of Dp8–23(Le et al. 1995). The positions of the “islands” of complex DNA (Tahiti, Moorea, Bora Bora, and Maupiti) are shown, as is the position of the new euchromatin/heterochromatin junction (“breakpoint”). The 420-kb functional centromere was localized by examining the transmission behavior of a large collection of minichromosome derivatives (Murphy and Karpen 1995b). Restriction mapping and hybridization analysis indicated that the centromere portion of γ1230 contained two satellite DNA blocks (AATAT and AAGAG), five small islands of complex DNA (Motus) inserted into the AATAT block, and a large region of complex DNA (Maupiti) at the right end (Sun et al. 1997). The sizes of the satellite blocks are shown.
To gain a more detailed understanding of the sequence composition and organization of the Dp1187 centromere, we used PFGE-purified γ1230 as an enriched source for cloning and sequencing. Heterochromatin contains a nonrandom distribution of restriction sites, as a result of the predominance of simple repeats. Therefore, purified γ1230 DNA was randomly sheared then cloned into the λ ZAPII vector (see Methods for details). The γ1230 library includes sequences from the centric heterochromatin, the subtelomeric heterochromatin (∼100 kb), and the euchromatin (∼100 kb). We screened the library with clones corresponding to the five major components previously demonstrated to be present in the centromere: the transposons, the AAGAG and AATAT satellite regions, the transposon-satellite junctions, and the complex DNA “island” Maupiti (Fig. 1; Sun et al. 1997). Previously unidentified types of DNA were isolated by randomly picking clones, and by probing the library with whole, gel-purified γ1230 and a specific restriction fragment from the centromeric region (see Methods). Plasmids were then generated by superinfection/excision of the λ clones. All clones were initially sequenced using the flanking T3 and T7 primers present in the pBluescript polylinker, and then sequences were extended with internal primers (see Methods). The positions of the clones within γ1230 were confirmed by hybridization to digested γ1230, using PFGE Southern analysis (data not shown).
We isolated, converted to plasmid, and sequenced 459 λ clones, and a total of 947.9 kb of sequence was generated from γ1230 (Table 1). This includes 495.6 kb of centromere sequence, which was assembled into 13 contigs (79.9 kb) that cover 19% of the 420 kb centromere. All complex DNA within the centromere (69.7 kb) has been sequenced using this library, and the average coverage for these regions was five- to sevenfold. In addition, the γ1230 library sequences include 10.2 kb (3%) of the satellite DNA, predominantly from the regions flanking the transposons (fivefold–sevenfold coverage) (see below). However, additional satellite sequence has been obtained by polymerase chain reaction (PCR) methods (see below). Most of the clones and sequence corresponded to subtelomeric, euchromatic, or unassigned regions of γ1230 (Table 1), and were not analyzed further.
Table 1.
Summary of Sequenced γ1230 Library Clones
| Region | Clones | Sequences | ||||
| no. | avg. size (kb) | total (kb)a | contig size (bp)b | type | sequence (bp)c | |
| Beagle (Motu 1) | 16 | 2.8 | 52.5 | 8043 | L AATAT | 726 |
| Beagle | 7063 | |||||
| R AATAT | 254 | |||||
| F (Motu 2) | 17 | 3.3 | 71.4 | 6431 | L AATAT | 942 |
| F | 4713 | |||||
| R AATAT | 776 | |||||
| 412 (Motu 3) | 32 | 2.7 | 82.4 | 9149 | L AATAT | 970 |
| 412 | 7572 | |||||
| R AATAT | 607 | |||||
| Bel (Motu 4) | 13 | 2.6 | 46.5 | 6888 | L AATAT | 582 |
| Bel | 6129 | |||||
| R AATAT | 177 | |||||
| You (Motu 5) | 5 | 1.7 | 24.7 | 7568 | L AATAT | 679 |
| 5333 | ||||||
| R AATAT | d950 | |||||
| R TTCTC | d606 | |||||
| Maupiti | 101 | 2.9 | 210.2 | 39,125 | L TTCTC | 201 |
| Maupiti | 38,924 | |||||
| Pure AATAT | 2 | 1.5 | 3.7 | e1058 | AATAT | e1058 |
| Pure TTCTC | 4 | 0.4 | 4.2 | f1627 | TTCTC | f1627 |
| CEN Subtotals | 190 | 2.8 | 495.6 | 79,889 | Complex | 69,734 |
| Satellite | 10,155 | |||||
| Breakpointg | 20 | 2.0 | 27 | 2655 | Euchromatin | 2291 |
| R AATAT | 364 | |||||
| Subtelomeric and Euchromatic | 35 | 2.4 | 47.3 | hNA | hNA | hNA |
| Unassigned | 214 | 3.0 | 378.0 | hNA | hNA | hNA |
| γ1230 Totals | 459 | 2.6 | 947.9 | 82,544 | Complex | 72,025 |
| Satellite | 10,519 | |||||
Total amount of sequence generated from all clones for each region.
Size of contig assembled for each region. L and R indicate position of the satellite to the left and right of the complex DNA (predominantly transposons), respectively (Fig. 2); see Figure 2 for transposon orientations.
Amount of satellite DNA sequence in the contig corresponding to the indicated junction.
At the 3′ end of the You contig, the transposon is juxtaposed to a 950-bp block of AATAT satellite, which in turn is juxtaposed with a block of TTCTC satellite.
Total for partial sequences from two clones, which both contained contigs of 529 bp.
Total for complete sequences from four clones, which contained contigs of 347, 439, 500, and 500 bp.
The breakpoint between the centromere and euchromatin produced during the generation of γ1230, see Figure 1. The AATAT satellite present in this clone marks the 5′ end of the centromere, but is not included in the CEN TOTALS for simplicity.
Assemblies of various sizes were produced, but are not included here because they are not relevant to centromere structure.
Single Transposons Are Intact and Nearly Identical to Previously Sequenced Elements, and Are Inserted Directly Into the AATAT Array
The γ1230 library clones have been used to generate complete sequences of the Motus, and have allowed us to characterize the junctions with the AATAT satellite (Table 1, Fig. 1). The Beagle, F, 412 and Bel transposons are intact and complete; all four elements have >99% identity with the reference transposon sequences deposited in GenBank. Motu 5 is >99% identical to the recently described transposon You, a non-LTR (LINE-like) retroposon (FlyBase 2002). All five transposons are inserted directly into the AATAT arrays, not into complex DNA, and are oriented as shown in Figure 2, confirming the orientations predicted from previous restriction mapping (Sun et al. 1997). The two long terminal repeats (LTRs) present in all three retrotransposons (Bel, Beagle, 412) are 99.7, 99.2, and 99.8% identical (respectively). Thus, these elements are likely to be recent insertion events (SanMiguel et al. 1998), or the sequences are under severe functional constraints. The completeness of the transposons and their recent origin raise questions about the evolution and function of these centromeric elements (see Discussion).
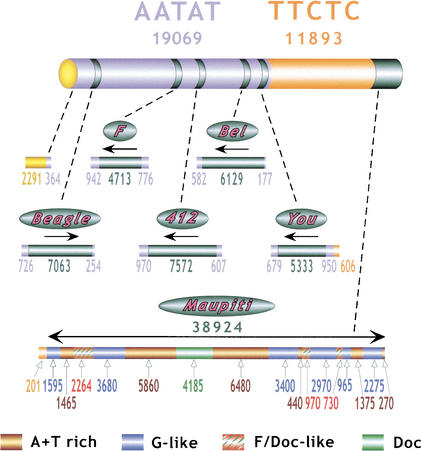
Figure 2.
Summary of γ1230 centromeric sequences. Numbers below AATAT and TTCTC report the total amount of sequence (base pairs) generated for each satellite, including blocks flanking complex DNA as well as the random, unmapped sequences generated by tagged PCR. Numbers below each transposon “bar” and the Maupiti diagram report the amount of contiguous sequence generated for each region and type of DNA, in base pairs. Arrows indicate the 5′ to 3′ orientation of the transposons, relative to previously sequenced euchromatic elements.
“Tagged” PCR Produces Sequence From Pure Satellite Arrays
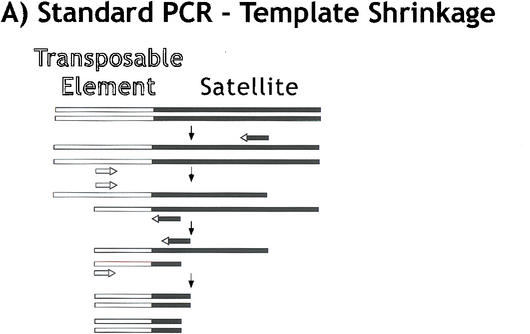
The analysis of sequences from the γ1230 library also demonstrated that the AATAT satellite is oriented as AATAT (from left to right in Fig. 1) at the euchromatic/centromere breakpoint and in the transposon flanks, and that the AAGAG satellite is oriented as TTCTC (designated as such hereafter). Analysis of the γ1230 library clones produced a total 7027 bp of AATAT contigs from the transposon flanks and euchromatin-heterochromatin junction, and 807 bp of contiguous TTCTC sequence from the AATAT-TTCTC and TTCTC-Maupiti junctions (Table 1). However, only six clones that contained pure satellite arrays were recovered from the library (Table 1). We have determined that the absence of pure satellite clones occurs during the initial cloning step into λ ZAP, and not in the subsequent excision step that generates the plasmid (data not shown). Because clones from the transposon-satellite junctions only contained up to 970 bp of satellite DNA, we suspect that clones containing satellite arrays larger than 1 kb are unstable, and that the lack of pure satellite clones reflects the 2-kb minimal size cutoff used to generate the library (see Methods).
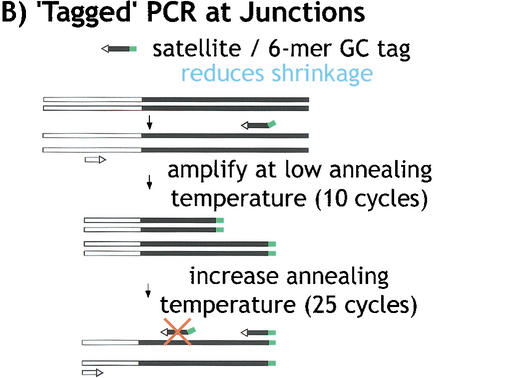
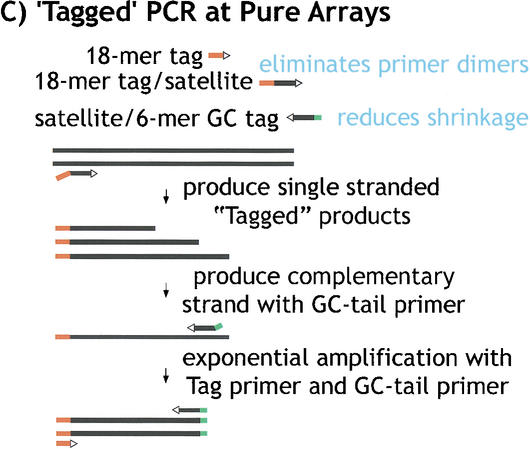
To further investigate the organization and composition of the satellite arrays, we developed PCR methods that would randomly sample satellite sequences. First, we generated random satellite clones and sequence using PCR amplification of gel-purified γ1230, with hybrid primers that contained satellite sequence plus a unique tag (Fig. 3). This approach was used to generate clones and sequences from the transposon-satellite junctions and from pure satellite arrays. One challenge in attempting to amplify junctions or pure satellite arrays is template shrinkage (Fig. 3A). We successfully ameliorated this problem by using hybrid satellite-tagged primers; transposon-satellite junctions required only one tagged primer plus one standard primer corresponding to the complex (transposon) DNA (Fig. 3B; supplemental material). Amplification and cloning of pure satellite arrays poses a problem in addition to template shrinkage—two primers that contain complementary satellite strands frequently form primer dimers, even when hybrid primers are used. Thus, to amplify pure satellite arrays, we used two different tagged primers and a primer corresponding to one of the tags (Fig. 3C). Second, we also generated pure satellite sequences using bacterial transposon insertion into gel-purified minichromosome DNA in vitro, followed by amplification with one primer from the bacterial transposon and a hybrid satellite-tagged primer (Fig. 3B).
Figure 3.
“Tagged” polymerase chain reaction (PCR) methods for cloning and sequencing heterochromatin. (A) Standard PCR amplifications with one satellite primer and one “unique” primer (homologous to the end of the transposon) result in shrinkage with each round of amplification. A satellite repeat primer will anneal anywhere in the repeated template, not just at the ends, and thus successive rounds of amplification will result in shorter and shorter products. The template shrinkage problem is even greater when two satellite primers are used to amplify pure satellite arrays (not shown), and in addition, the self-complementarity of the two satellite primers results in primer-dimer formation (not shown). (B) Junctions between complex DNA (transposons in this case) and satellite arrays are successfully amplified with significantly less shrinkage when a hybrid GC-tagged/satellite primer is used. Initial amplification at low stringency incorporates the tag at random within the satellite array then extends across the location of the TE primer; the subsequent exponential amplification reduces shrinkage because high stringency mandates annealing of the entire hybrid primer. This method was also used to generate sequence from bacterial transposon insertions into gel-purified γ1230; in this case, the “complex” primer was homologous to the end of the bacterial transposon. (C) Pure satellite arrays were amplified with two tagged primers, plus a primer corresponding to one of the tags. One tagged primer was used for single-stranded extension at low stringency; after gel isolation of the single-strand products, the second primer was used to synthesize the complementary strand, then the tag and one tagged primer were used for high stringency amplification.
The tagged PCR experiments produced significant coverage of both pure satellite arrays and junctions with complex sequences (Table 2). The PCR sequences increased the coverage for the AATAT that flanks the single transposons to >10-fold, from the fivefold–sevenfold coverage produced by the γ1230 library. Genome Priming System (GPS) insertion into gel-purified γ1230, using one tagged satellite primer and one primer homologous to the bacterial transposon (Fig. 3B), produced 11 kb of pure AATAT sequence. This total does not include 3 kb of shorter AATAT sequences (<200 bp) that were identical to larger PCR product sequences. We conservatively chose to exclude these sequences from totals presented here and in the tables, as they could represent shrinkage products from the same sites as the larger products. PCR with two tagged TTCTC primers (Fig. 3C) generated 6.4 kb of pure satellite sequence, and GPS insertion produced 3 kb of TTCTC sequence. However, we recovered no clones using two tagged AATAT primers (Fig. 3C). This result was likely caused by primer-dimer formation that occurs at the low annealing temperature that had to be used with the AATAT primer. Regardless, these results demonstrate the utility of the tagged PCR and transposon insertion approaches to analyzing satellite DNA, methods that can be used to analyze similar regions in the Drosophila and human genomes.
Table 2.
Summary of Satellite DNA Sequences from Different Sources
| Satellite | Source | No. of clones sequenced | Sequence (bp)a | Average length (bp) | Longest (bp)b |
| AATAT | Library | 13b | 8085 | 622 | 970 |
| Tagged PCR | 0 | — | — | — | |
| GPS Insertion | 76 | 10,984 | 146 | 432 | |
| Subtotal | 89 | 19,069 | 214 | 970 | |
| TTCTC | Library | 6 | 2434 | 406 | 606 |
| Tagged PCR | 40 | 6432 | 161 | 455 | |
| GPS Insertion | 27 | 3027 | 112 | 203 | |
| Subtotal | 73 | 11,893 | 163 | 606 | |
| Totals | 162 | 30,962 | 191 | 970 |
Total bp in contigs.
For the purposes of this summary, each transposon-AATAT junction was considered to be one clone.
To estimate the error rate in the γ1230 sequences, we compared the sequences derived from the γ1230 library λ clones to the homologous PCR-generated sequences. The transposon and satellite components of the junction fragments were 100% identical in the λ and PCR-derived consensus sequences. The depth of coverage allowed us to eliminate errors generated by sequencing, PCR, or cloning. Differences of only 0.1% were observed when comparing individual reads from the GPS transposon sequences in the clones that were generated after insertion into isolated γ1230, and with the published transposon sequence. Similarly, comparison of the flanking satellite DNA sequence with the consensus sequence derived from either the PCR and λ sequences only showed a 0.1% difference. Importantly, in almost all cases where there was a base pair ambiguity, only one sequenced strand was different; the remaining sequences were in complete agreement. This gives us a valid prediction for the error rate present in the pure satellite sequences. The observed differences of 0.1% are roughly fourfold higher than what we would expect from previously determined TAQ polymerase error rates (Cline et al. 1996). This error in some of the PCR clones could be from the result of the insertion process (gene conversion), two rounds of PCR (isolation and screening), sequencing error, or different PCR conditions. It is likely that most of the observed differences among individual reads were generated during PCR or cloning, rather than sequencing. Thus, the quality of the sequences presented here is high, and in most cases the depth of coverage allowed us to generate consensus sequences with high confidence.
The AATAT and TTCTC Satellite Arrays Are Highly Conserved and Uniform
Tables 2 and 3, and Figure 2 summarize the satellite sequence totals and characteristics obtained from analysis of the library and the amplified products. In total, 31 kb of satellite DNA sequence was obtained from analysis of the γ1230 library and the PCR approaches, corresponding to ∼8.7% and 7.5% of the AATAT and TTCTC satellites present in the centromere, respectively. The small size of the clones allowed us to generate complete sequences using vector primers (see below). The validity of the assembled sequences was supported by the fact that consensus sequence information obtained from the λ clones and the PCR approaches for the transposons and satellite flanks are identical (differences for individual reads = 0.1%, see above).
Table 3.
Summary of Satellite DNA Sequence Variations
| Satellite/source | Sequence | Changes | Types of changes# (%)d | ||||||||||
| bpa | %b | # | %c | ||||||||||
| AATAT | A → T | A → G | A → C | T → A | T → G | T → C | Ins/del | TS | TV | ||||
| Junctionse | 7027 | 3.2 | 180 | 2.6 | 48 (27) | 43 (24) | 5 (3) | 55 (31) | 0 | 6 (3) | 23 (13) | 49 (31) | 108 (69) |
| Pure | 12,042 | 5.5 | 233 | 1.9 | 59 (25) | 59 (25) | 5 (2) | 75 (32) | 3 (1) | 12 (5) | 20 (9) | 71 (33) | 142 (67) |
| Total | 19,069 | 8.7 | 413 | 2.2 | 107 (26) | 102 (25) | 10 (2.5) | 130 (32) | 3 (0.7) | 18 (4.5) | 43 (10) | 120 (32) | 250 (68) |
| TTCTC | T → A | T → G | T → C | C → A | C → T | C → G | Ins/del | TS | TV | ||||
| Junctionsf | 807 | 0.1 | 10 | 1.2 | 1 (10) | 0 | 8 (80) | 1 (10) | 0 | 0 | 0 | 8 (80) | 2 (20) |
| Pure | 11,086 | 7.4 | 21 | 0.2 | 1 (5) | 1 (5) | 11 (52) | 0 | 4 (19) | 0 | 4 (19) | 15 (88) | 2 (12) |
| Total | 11,893 | 7.5 | 31 | 0.3 | 2 (6.5) | 1 (3.2) | 19 (61) | 1 (3.2) | 4 (13) | 0 | 4 (13) | 23 (85) | 4 (15) |
Total bp in contigs
% of total satellite in γ1230, see Figure 1
% of total sequence changed
% of all changes; TS = transitions (C ↔ T, A ↔ G), TV = transversions (A ↔ T, T ↔ G, C ↔ G, C ↔ A) (TS and TV summary percentages exclude insertions/deletions)
Junctions between transposons and AATAT, and AATAT-TTCTC.
Junctions between AATAT-TTCTC and TTCTC-Maupiti.
These approaches have produced the largest data set of Drosophila satellite sequences to date, providing an opportunity to perform a detailed analysis of the sequence and organization of the satellite arrays. First, tandem repeats within satellite arrays are oriented in the same direction (“head to tail”), AATAT and TTCTC (left to right, Fig. 1). Second, an AATAT/TTCTC junction is located 950 bp from the right end of the You element. Thus, the two satellites appear to be directly juxtaposed, with no intervening complex DNA, or other type of repeat. Third, the arrays are uniform in sequence; only 2.2% and 0.3% of the AATAT and TTCTC sequences contained base changes, respectively. The vast majority of alterations (92%) were one base changes to the simple sequences, and there were no significant insertions of complex DNA (besides the transposons in the Motus). The remaining changes were small insertions and deletions (<5 bases), which could also represent multiple single-base changes. Only a small proportion of the observed AATAT variation results from mutation during PCR amplification or cloning or sequencing errors (expect 0.1% variation, see above). The absence of significant variability across the bulk of the sequenced satellites is consistent with the high resolution restriction mapping (Sun et al. 1997), and demonstrates that the satellite arrays are uniform and lack significant insertions of complex DNA (besides the transposons in AATAT).
Important information about the composition of the satellite arrays was revealed by analyzing the frequencies of different types of nucleotide changes (Table 3). First, TTCTC displayed significantly fewer sequence changes than AATAT (0.3% versus 2.2%, χ2 = 124, P < .0001). Second, the AATAT arrays at the junctions displayed nearly identical frequencies of different types of changes as the “pure” arrays (Table 3). The only exception was a moderate but statistically insignificant increase in insertions/deletions near the junctions, from 9% to 13% (χ2 = 0.52, P < .50). Thus, transposon insertions did not alter the frequencies of changes in the flanking satellite or the expansion of altered repeats. Third, the overall frequency of transversions (68%) was twofold greater than transitions in both pure AATAT arrays and junctions (32%; χ2 = 19, P < .0001; see Discussion). This ratio was reversed for the TTCTC satellite; however, the significance of this result is unclear, as only 27 single base alterations were identified in these arrays. Finally, we observed clustering of polymorphisms and regular spacing (5 and 10 bp) of units with the same base changes (data not shown), perhaps reflecting expansion of individual altered repeats during array evolution. In summary, we conclude that the sequence and organization of the AATAT and TTCTC arrays are highly conserved, and that the two types of satellites display different frequencies and types of base changes.
Assembly and Analysis of the Maupiti Region
One of the greatest challenges we faced was assembling a complete contig of the complex island Maupiti. The high-resolution restriction map suggested that this island was ∼35 kb in size, and contained multiple, partial G-like transposons, an incomplete Doc transposon, and at least two large blocks of a novel A+T-rich sequence (Sun et al. 1997). The large size and repetitive nature of this region created significant difficulties for sequence assembly. Existing alignment/assembly programs are not able to automatically assemble repeated sequences, even if the repeats are variable; sequences are frequently misplaced within the contig. We were able to overcome these difficulties by isolating multiple clones from the λ library for each region, which confirmed the presence and validity of polymorphisms that were necessary to correct assembly. Large clones (e.g., 4–7 kb) that spanned multiple polymorphisms and components were especially helpful in validating the assembly of smaller contigs. In vitro insertion of a modified yeast transposon (AT-2 [Devine and Boeke 1994], see Methods) was used to complete the sequences of plasmids that were difficult to assemble because of the presence of repetitive DNA. Finally, Sequencher 3.0 software (GeneCodes) was helpful, because it allowed us to manually align the most difficult sequences, and to simultaneously view the sequence traces to confirm polymorphisms. Other regions of heterochromatin (e.g., pure satellite arrays with few polymorphisms) will likely be even more challenging, but assembly of the Maupiti contig has served as an important stepping stone and learning experience that will help us design strategies to assemble even more challenging heterochromatic sequences.
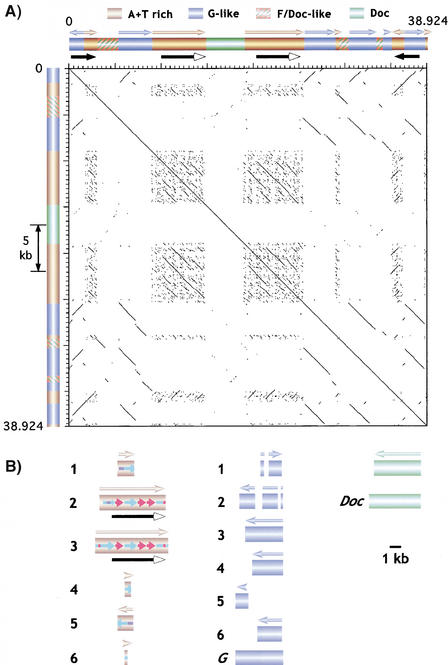
The assembled Maupiti contig (Fig. 2) was based on an average of fivefold–sevenfold coverage, and revealed information about this region of heterochromatin that was not visible in the high-resolution map. The size of Maupiti is 38.924 kb (Table 1, Fig. 2), ∼10% larger than the 35 kb estimated from the restriction map (Sun et al. 1997). Maupiti contains only four types of sequences: 16.2 kb of A+T-rich sequence (Sun et al. 1997), 15.1 kb of G-like transposon sequence, 4.3 kb of F/Doc-like transposon sequence, and 4.2 kb of a Doc transposon. This analysis also identified the junction between the TTCTC array and the left end of Maupiti, which demonstrated that the orientation of this satellite is the same at the left and right ends of the block (Fig. 1).
The assembled Maupiti contig revealed the exact size, distribution, and sequence of the A+T-rich DNA, previously identified in a small clone in the Sun et al. study (1997). The A+T-rich sequence is present in six blocks of 1550, 5900, 6500, 550, 1450, and 270 bp, from left to right (brown cylinders, Fig. 2). These blocks share between 88% and 100% identity, predominantly in the 95–98% range. All blocks except block number five are oriented in the same direction. The dot plot (Fig. 4A) and sequence comparisons (Fig. 4B; see Methods) demonstrate that the A+T-rich blocks contain related, small tandem repeats (small colored arrows, Fig. 4B). The largest blocks (repeats 2 and 3) are nearly identical; each contains a 4.65-kb direct repeat, plus different combinations of subrepeats in the left ends. The smaller blocks contain different combinations of these subrepeats. Blocks 1 and 5 are identical, but are in opposite orientation (see below).
Figure 4.
Sequence composition and organization of Maupiti. (A) Dot-plot analysis of Maupiti reveals the presence and organization of internally repeated DNAs. Arrows above each bar indicate relative orientations of each component. Solid arrows below the bars indicate the presence of inverted repeats at the ends, and large internal direct repeats. Note that most of the internal elements are oriented in the same direction. (B) The substructures of the A+T-rich, G-like, and Doc elements are shown relative to each other, and to previously sequenced elements (bottom). Blocks are numbered from left to right (relative to A). The homologies (colors) and relative orientations (arrowheads) of subrepeats within the A+T-rich elements are shown. The inverted repeats at the ends are composed of G-like block 1/A+T-rich block 1 and A+T-rich block 5/G-like block 6.
The assembled Maupiti sequence also revealed the positions and orientations of the transposons (Fig. 4). All the transposons in Maupiti are incomplete or fragmented, unlike the complete transposons in the AATAT Motus (Fig. 2). The Doc element is 98% identical to sequenced euchromatic Doc elements, and is nearly complete (green, Fig. 4). The F/Doc elements (red with green cross-hatch, Fig. 4) represent fragments that are 80%–87% identical to sequenced euchromatic elements (parts of F and Doc elements share significant homology). These elements appear to be a subfamily that shares more sequence homology with each other than with previously sequenced F and Doc elements reported in GenBank. A similar conclusion can be drawn from analysis of the G-like elements. The six blocks (blue cylinders, Fig. 4B) are 91%–100% homologous to each other, and share 80%–89% identity with parts of previously sequenced G elements. None of these G-likeelements are complete, and all copies are oriented in the same direction, except block 1 (Fig. 4B). Thus, these elements appear to be a subfamily of G-like repeats that are more closely related to each other than to previously sequenced G elements. These observations suggest that local repeat expansion played an important role in generating the structure of Maupiti (see Discussion). Finally, identical 3-kb blocks composed of G-like and A+T-rich sequences are present at both ends, in inverted orientation (Fig. 4A).
DISCUSSION
Successful Methods for Sequence Analysis of Centric Heterochromatin
Current genome projects have not produced truly complete genome sequences; the human and Drosophila assemblies lack the ∼30% of these genomes considered to be heterochromatic. Heterochromatin sequence and organization has been notoriously difficult to analyze, as a result of the presence of repeated DNA. Heterochromatin mapping, cloning, and sequencing requires approaches that are not necessary in the analysis of euchromatic, predominantly single or low copy number DNA (Schueler et al. 2001). One approach is to use methods that isolate specific regions of heterochromatin from the rest of the genome. Sequences that are repeated many times in the genome are often present in only one or a few copies in a particular region, which reduces the complexity of mapping, sequencing, and assembly. Single-copy entry points, in the form of marked transposon insertions (e.g., P elements in Drosophila), or rearrangements that juxtapose euchromatin with heterochromatin, have been used successfully for heterochromatin genome analysis (Karpen and Spradling 1990; Karpen and Spradling 1992; Howe et al. 1995; Le et al. 1995; Sun et al. 1997; Yan et al. 2002).
In this study, PFGE isolation of the γ1230 minichromosome derivative provided a purified template for cloning and PCR, which allowed us to clone, sequence, and assemble a significant proportion of the repeat-rich, 420-kb functional centromere. The entire complex DNA component of the minichromosome centromere (69.7 kb) was sequenced and assembled, including Maupiti, and the five transposons inserted in the AATAT array. Analysis of γ1230 library clones produced 10.5 kb of satellite sequence, predominantly from the AATAT arrays that flank the transposons. Tagged PCR and GPS transposon insertion using purified γ1230 were very successful at producing satellite DNA sequence from both pure arrays and junctions. From all methods, 19.1 kb of AATAT sequence and 11.9 kb of TTCTC sequence were generated, corresponding to >8% of the amount of each satellite in the minichromosome centromere (Table 2). The sequence coverage averaged fivefold–sevenfold for the transposons and Maupiti, and >10-fold for the AATAT flanking the single transposons. Comparison of sequences derived from λ cloning to the PCR-generated sequences demonstrated that they differed by only 0.1%, which suggests that the quality of the sequence is high. We conclude that heterochromatin can be successfully analyzed using these modifications of standard genomic approaches. One useful application of these approaches will be in the analysis of other heterochromatic regions that are isolated from “contaminating” genomic background sequences, for example the heterochromatic Bacterial Artificial Chromosome (BAC) clones generated by the Drosophila, human, and Arabidopsis genome projects.
Sequence Composition and Organization of the Dp1187 Centromere
Although the satellite arrays have not been completely sequenced, this study has produced the largest collection of Drosophila satellite sequences to date (31 kb), providing an opportunity to gain a greater understanding of the composition and organization of Drosophila satellites. We found that the AATAT and TTCTC components of the Dp1187 centromere are organized as tandem repeats in a uniform “head-to-tail” orientation, with few examples of “head-to-head” or “tail-to-tail” orientations. The arrays are oriented as AATAT and TTCTC, from left to right (Figs. 1 and 2), at all sites where orientation could be determined unambiguously (i.e., in the transposon flanks, the AATAT/TTCTC junction, and the Maupiti junction). We also determined that the AATAT and TTCTC arrays are directly juxtaposed 950 bp from the right end of the You element, with no intervening sequences.
The level of sequence variation among repeats was low; only 2.2% of the bases in the AATAT array were altered, consisting of predominantly single base insertions or deletions. This is similar to the level of variation observed in comparisons of different human X chromosome α satellite arrays (1%) (Durfy and Willard 1989), and in interspecies comparisons of the PRAT satellite family in Coleoptera (1.3%) (Mravinac et al. 2002). In contrast, only 0.3% of the TTCTC bases were altered. The TTCTC array could be newer than the AATAT array, and thus may not have had as much time to accumulate base alterations. Alternatively, molecular-drive mechanisms (Dover 1982), for example array expansion and contraction resulting from unequal exchange, and repeat homogenization through gene conversion, may act differently on the two sequences. It is also possible that TTCTC contains fewer variations from functional constraints, such as sequence requirements that facilitate centromere function (see below). Unfortunately, there are no published, large-scale compilations of naturally occurring sequence variations for Drosophila heterochromatin or euchromatin. Thus, the generality of these findings, and their relevance to the evolution and function of these satellite arrays in Drosophila require further detailed analysis of naturally occurring variations in different satellites, and in similar satellite sequences located in different sites in the genome. Such studies may also help distinguish between the molecular mechanisms that are responsible for satellite evolution in Drosophila.
The pattern of base alterations in the satellite arrays was unusual. Approximately two-thirds of the AATAT changes were transversions, and only one third were transitions. This result is surprising, as transitions are thought to be significantly more frequent than transversions. The predominance of transversions could be from the molecular characteristics of the mechanisms responsible for mutation and homogenization, and how they act on this particular sequence, or could result from functional selection. Again, additional large-scale studies of natural variation in Drosophila and other species are necessary to determine if the high frequency of transversions occurs in other satellite arrays, and if heterochromatin and euchromatin display similar frequencies of transversions and transitions.
We generated complete sequences for the five transposons inserted into the AATAT array. Heterochromatic transposons previously identified in Drosophila heterochromatin are often organized as scrambled clusters of different, incomplete elements (Devlin et al. 1990; Trapitz et al. 1992; Hochstenbach et al. 1994). Surprisingly, all five elements in the Dp1187 centromere are intact and complete, in comparison to sequenced euchromatic elements. The two LTRs in the Bel, Beagle, and 412 elements displayed very high levels of cis homology (99.2–99.8% identity). Interestingly, these transposons are inserted directly into the AATAT arrays, with no insertion of other sequences at the junctions. In fact, the satellites present at the junctions display similar levels of variation in comparison to the “pure” satellite arrays obtained by random sampling (Table 3). The completeness of the transposons, the LTR homologies, and the lack of AATAT divergence at the junctions suggest that these elements were recent additions to the array (SanMiguel et al. 1998). Alternatively, the transposon sequences may be constrained by functional requirements, such as centromere activity. Recent studies have demonstrated that centromeric silencing in Schizosaccharomyces pombe and DNA elimination in Tetrahymena thermophila require RNA interference (RNAi) mechanisms, which act on repeats that apparently evolved from transposons (for review, see Dernburg and Karpen 2002). Finally, no transposons were found in the TTCTC array; this is another piece of evidence in favor of a more recent origin of this array, relative to AATAT (see above), but could also reflect insertion sequence bias of the transposons. It will be interesting to determine if other TTCTC arrays lack transposons, and whether other AATAT arrays contain single, intact, presumably recent insertions.
The overall organization and sequence composition of the 39 kb Maupiti contig provides some clues concerning its molecular evolution. The ends, consisting of A+T-rich and G-like sequences, are in inverted orientation (Fig. 4A). However, the elements present in the rest of the contig are organized as direct repeats, even though they are separated by other components. This observation, combined with the high homology of the different blocks, suggest that Maupiti may have evolved by successive duplications, unequal exchanges, and partial deletions. The basic building block may have been an A+T-rich unit, which served as a target for insertion of a G-like element, followed by rounds of expansion and deletion. The substructure of the A+T-rich subrepeats also suggests duplication of a basic subunit and evolution via similar molecular drive mechanisms (Dover 1982). Finally, the nearly complete Doc element is present in only one copy and may have inserted into the repeated array subsequent to the repeat duplications.
What Is the Role of DNA Sequence in Centromere Identity?
Analysis of the sequence characteristics of centromeres in different organisms, and the fact that centromeric satellites are neither necessary nor sufficient for centromere function, suggest that primary DNA sequence does not determine centromere identity and propagation (see Introduction, and Choo 2000; Sullivan et al. 2001). Sampling of 8.4% of the satellite arrays in the Dp1187 centromere demonstrated that they are uniform, and do not contain any blocks of complex DNAs. Furthermore, the transposons inserted in the AATAT array are nearly identical to elements present at many sites within the euchromatin and the heterochromatin, and Maupiti does not contain unique, centromere-specific sequences. These results confirm our previous assertion that the Dp1187 centromere does not contain sequences that are centromere-specific, or are located at all Drosophila centromeres (Murphy and Karpen 1995b; Sun et al. 1997). There is significant additional evidence in favor of epigenetic models for the propagation of centromere identity mechanisms, which can parsimoniously account for both centromere stability and plasticity (see Introduction and Karpen and Allshire 1997; Sullivan et al. 2001).
If epigenetic mechanisms determine centromere identity and function, do secondary sequence characteristics play a role, such as repetition and sequence composition, or the ability to form bent structures (Vig 1994; Murphy and Karpen 1998; Koch 2000; Sullivan et al. 2001)? All sequenced endogenous eukaryotic centromeres are A+T-rich, and contain repeated DNA, usually in the form of simple or complex satellite DNA. This study demonstrates that the minichromosome centromere is also A+T-rich and replete in highly repeated satellite DNA. However, neocentromeres in humans are established on normal euchromatic DNA, which does not share these properties, or any obvious conserved motifs (Barry et al. 2000; Lo et al. 2001a,b; Satinover et al. 2001).
Higher-order organization of centromeric DNA, such as the inverted repeat structure at the ends of Maupiti, may be important for function. All three S. pombe centromeres contain transposon-like, middle-repetitive elements that flank a single copy central core in an inverted orientation (Takahashi et al. 1992; Clarke et al. 1993). Interestingly, the ends of Maupiti are also organized as inverted repeats. It has been hypothesized that inverted repeats may cause centromeric chromatin to assemble a stem-loop structure in S. pombe, and similar types of structures have been proposed for flies and mammals (Sullivan et al. 2001; Blower et al. 2002). Centromeric chromatin in fly and mammalian metaphase chromosomes exhibit unique, cylindrical structures, which may rely on the presence of repeated DNA, inverted repeats, or some other characteristic of the sequence, such as A+T composition (Blower et al. 2002). This higher-order structure may be necessary to “present” the centromeric chromatin to the outside of the condensed chromosome, ensuring its contacts with components responsible for kinetochore formation and interactions with microtubules. Further analysis is necessary to determine if any property of centromeric sequences affects formation of this structure or other aspects of centromere structure and function.
How Can We Generate Truly Complete Genome Sequence Assemblies?
We and others (e.g. Schueler et al. 2001) have successfully demonstrated that the difficulties associated with analyzing the sequence composition of heterochromatin, including satellite DNA, can be overcome using modifications of extant genomic methods. Can the methods described here be used to produce truly complete genome sequences? Many of these methods, such as gel-purification and cloning of heterochromatic DNA and tagged PCR, can be applied to studies of other regions of heterochromatin in flies and other organisms. However, these approaches are not efficient or robust enough to be scaled up to analyze the ∼100 Mb of heterochromatin in the Drosophila genome (Adams et al. 2000). Complementary methods for focusing on specific regions of heterochromatin involve generating new, single-copy entry points. Chromosome rearrangements that juxtapose single-copy euchromatic regions with heterochromatin have provided a method for long-range restriction mapping and gene localization, allowing the single-copy region to be used as a probe to tag one end of the restriction fragments (Karpen and Spradling 1990; Howe et al. 1995; Le et al. 1995). Single P-transposable elements provide another method for marking and analyzing specific heterochromatic regions (Karpen and Spradling 1992; Thompson-Stewart et al. 1994; Howe et al. 1995; Roseman et al. 1995; Wallrath and Elgin 1995). We have generated over 600 single P-element insertions into Drosophila centric heterochromatin, which can now be used to map, clone, sequence, and assemble many different regions (A. Konev et al., in prep; Yan et al. 2002).
Most importantly, even if significant amounts of heterochromatic sequence are generated, the manual approach used to assemble Maupiti is too inefficient. We need to develop automated assembly software that can appropriately join regions rich in repeated sequence. Regardless, our study demonstrates that one key to assembling repeated sequence is to focus on small regions of heterochromatin. Sequences repeated throughout the heterochromatin are often present “locally” in only one or a few copies. Focus on specific heterochromatic regions can thus lead to accurate contig assemblies, as we have shown for Maupiti. The constrained assembler used to assemble Drosophila and human whole-genome shotgun (WGS) sequences could be very useful for heterochromatin sequence assemblies, because retention of “mated-pair” information can overcome some of the misalignment problems caused by repeats (Myers et al. 2000). The manual Maupiti assembly we have generated can provide a good test case for the abilities of new assemblers to work successfully with repeated DNA.
In conclusion, a truly complete understanding of genome organization and sequence requires much more extensive mapping and sequencing of heterochromatin, in both model organisms and humans. This study demonstrates that significant amounts of heterochromatin sequence can be generated and assembled, and that important information about heterochromatin structure and biology can be produced by such an analysis. However, a large-scale genomics attack on heterochromatin will require development of high-throughput approaches, especially in the area of assembly software development.
METHODS
Drosophila Strains
The generation and gross structural analysis of γ1230 are described in Figure 1 and in (Le et al. 1995; Murphy and Karpen 1995b; Sun et al. 1997). The genotype for the strain used in all experiments was y; ry506; Dpγ1230, y- ry+. Standard culture conditions and media were used.
Library Construction and Screening
PFGE purified γ1230 DNA (Sun et al. 1997) was used to generate small insert clones for sequence analysis. Briefly, γ1230 DNA was randomly sheared to 2–7 kb, to avoid restriction-site biases that would preclude inclusion of many heterochromatic regions. The sheared DNA was blunted, EcoRI methylated, ligated to EcoRI linkers, digested with EcoRI, and fragments <1 kb were removed using a Sephacryl-drip column. The 2–7 kb fractions were then ligated to the EcoRI digested and dephosphorylated λ ZAPII vector. Characterization of clone numbers and insert size demonstrated that γ1230 was represented >80-fold. A high-quality, random-sheared library from total Drosophila genomic DNA was generated at the same time, with the same protocol; this library may be useful for analyses of other Drosophila heterochromatin regions.
The library was screened by hybridization to colonies. Probes were generated using information from the γ1230 restriction map (Sun et al. 1997), including all the previously identified transposons, the A+T-rich sequence, the AATAT and TTCTC satellites, gel purified, intact γ1230, and a γ1230 centromeric restriction fragment (235 kb SpeI). Approximately 100 colonies were also picked randomly and analyzed. The locations of clones within γ1230 (Sun et al. 1997) were then determined by PFGE Southern analysis.
Cloning Satellite DNA From Gel-Purified γ1230 by “Tagged” PCR and Bacterial Transposon Insertion
γ1230 DNA Template Preparation
To eliminate background from other genomic regions with similar repeats, γ1230 was gel-isolated away from genomic DNA using PFGE (Sun et al. 1997). Isolated γ1230 DNA was cut with NotI and SfiI restriction enzymes resulting in a 490-kb fragment (420-kb heterochromatin/centromere + 70-kb euchromatin). The 490-kb fragment was gel-isolated and treated with β-agarose to produce a suitable template for PCR.
Satellite “Tagged” PCR for Junctions
To reduce product shrinkage resulting from primers annealing to proximal repeat units, PCR primers composed of six units of AATAT or five units of TTCTC were tagged at the 5′ end with a high Tm, short 6 mer sequence (CCCGGG or GC GCGC). PCR amplification starting with ∼0.1 ng of DNA was carried out in 10 mM Tris-HCL (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.001% (w/v) gelatin, 200 μM of each dNTP, and 200 ng of each primer, in a 50-μL volume. PCR was performed under the following conditions: 94°C for 2 min, then 2–10 cycles of 94°C for 15 sec, 36°C for 15 sec, 72°C for 1 min (slow ramping 0.6°C/sec); 30 cycles of 94°C for 15 sec, 56°C for 30 sec, 72°C for 1 min; and a final extension time of 72°C for 8 min. Increasing the annealing temperature takes advantage of the 5′ primer tags that were incorporated in the initial cycles. This results in an increase of the product size from 50–75 bp of satellite flank to 500–600 bp of satellite flank (see supplemental materials). PCR products were cloned using PCR-Script Amp cloning kit (Stratagene) or TOPO TA kit (Invitrogen). Colonies were screened with colony PCR using M13 Rev and Fwd primers for pCR2.1 TOPO clones, and T3 and T7 primers for pPCR-Script AMP clones. PCR products were visualized on 2% Metaphor agarose gels and then clones containing inserts were purified using either QIAquick PCR Purification Kit (QIAGEN) or Wizard PCR Preps DNA Purification System (Promega). The same primers were used for both amplification and DNA sequencing. Sequences were analyzed with Sequencher (Genecodes).
PCR on Satellite Arrays
Amplification of satellite DNA from γ1230 was done by using a single satellite primer (five units of TTCTC) that was tagged at the 5′ end with an 18-mer sequence (5′-CATGACTGGAC GCTCCAC-3′ or 5′-GAGTTGCATCTGGGATCG-3′). PCR was performed under the following conditions: 94°C for 2 min; 30 cycles of 94°C for 15 sec, 36°C for 15 sec, (slow ramping 0.4°C/sec) 72°C for 2 min, one final extension of 72°C for 8 min. This produced single-stranded satellite sequences that incorporated the 5′ tagged 18-bp primer. After cleanup of the PCR reaction, 1–2 μL of the reaction was used as a template for PCR with the 18-mer and the junction primers. Amplification, isolation, and sequencing were the same as for the junctions.
Transposon Tagging
The Genome Priming System (NEB) was used according to manufacturer's instructions to introduce a modified bacterial transposon into the purified and isolated γ1230 DNA. The flanking satellite DNA was amplified using a primer from the end of the transposon and a satellite-tagged primer. The products were cloned and analyzed in the same manner for the satellite-tagged PCR method.
Sequence Determination and Analysis
All sequencing was performed in The Salk Institute DNA Sequencing Facility, using big dye-termination reagents and ABI/PE 377 automated sequencers. Sequencer 3.0 (Genecodes) was used for all sequence assemblies. Accession numbers are included in supplemental materials.
Acknowledgments
We thank Michael Blower, Patrick Heun, and Beth Sullivan for critical editorial comments. This research was supported by the National Institutes of Health/National Human Genome Research Institute (R01 HG00747).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL karpen@salk.edu; FAX (858) 622-0417.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.681703.
REFERENCES
- 1.The Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. In Nature, pp. 796–815. [DOI] [PubMed]
- 2.Adams M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- 3.Allshire R.C., Javerzat, J.P., Redhead, N.J., and Cranston, G. 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157-169. [DOI] [PubMed] [Google Scholar]
- 4.Arn P.H., Li, X., Smith, C., Hsu, M., Schwartz, D.C., and Jabs, E.W. 1991. Analysis of DNA restriction fragments greater than 5.7 Mb in size from the centromeric region of human chromosomes. Mamm. Genome 1: 249-254. [DOI] [PubMed] [Google Scholar]
- 5.Barry A.E., Bateman, M., Howman, E.V., Cancilla, M.R., Tainton, K.M., Irvine, D.V., Saffery, R., and Choo, K.H. 2000. The 10q25 neocentromere and its inactive progenitor have identical primary nucleotide sequence: Further evidence for epigenetic modification. Genome Res. 10: 832-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539-2542. [DOI] [PubMed] [Google Scholar]
- 7.Blower M.D., Sullivan, B.A., and Karpen, G.H. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2: 319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. In Science, pp. 2012–2018. [DOI] [PubMed]
- 9.Choo K.H. 2000. Centromerization. Trends Cell. Biol. 10: 182-188. [DOI] [PubMed] [Google Scholar]
- 10.Clarke L., Baum, M., Marschall, L.G., Ngan, V.K., and Steiner, N.C. 1993. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harbor Symp. Quant. Biol. 58: 687-695. [DOI] [PubMed] [Google Scholar]
- 11.Cline J., Braman, J.C., and Hogrefe, H.H. 1996. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24: 3546-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copenhaver G.P., Nickel, K., Kuromori, T., Benito, M.I., Kaul, S., Lin, X., Bevan, M., Murphy, G., Harris, B., Parnell, L.D., et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468-2474. [DOI] [PubMed] [Google Scholar]
- 13.Dernburg A.F. and Karpen, G.H. 2002. A chromosome RNAissance. Cell 111: 159-162. [DOI] [PubMed] [Google Scholar]
- 14.Dernburg A.F., Sedat, J.W., and Hawley, R.S. 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135-146. [DOI] [PubMed] [Google Scholar]
- 15.Devine S.E. and Boeke, J.D. 1994. Efficient integration of artificial transposons into plasmid targets in vitro: A useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 22: 3765-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devlin R.H., Holm, D.G., Morin, K.R., and Honda, B.M. 1990. Identifying a single-copy DNA sequence associated with the expression of a heterochromatic gene, the light locus of Drosophila melanogaster. Genome 33: 405-415. [DOI] [PubMed] [Google Scholar]
- 17.Dobie K.W., Hari, K.L., Maggert, K.A., and Karpen, G.H. 1999. Centromere proteins and chromosome inheritance: A complex affair. Curr. Opin. Genet. Dev. 9: 206-217. [DOI] [PubMed] [Google Scholar]
- 18.Dobie K.W., Kennedy, C.D., Velasco, V.M., McGrath, T.L., Weko, J., Patterson, R.W., and Karpen, G.H. 2001. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics 157: 1623-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaldson K.M., Lui, A., and Karpen, G.H. 2002. Modifiers of terminal deficiency-associated position effect variegation in Drosophila. Genetics 160: 995-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dover G. 1982. Molecular drive: A cohesive mode of species evolution. Nature 299: 111-117. [DOI] [PubMed] [Google Scholar]
- 21.Dunham I., Lengauer, C., Cremer, T., and Featherstone, T. 1992. Rapid generation of chromosome-specific alphoid DNA probes using the polymerase chain reaction. Hum. Genet. 88: 457-462. [DOI] [PubMed] [Google Scholar]
- 22.Durfy S.J. and Willard, H.F. 1989. Patterns of intra- and interarray sequence variation in α satellite from the human X chromosome: Evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics 5: 810-821. [DOI] [PubMed] [Google Scholar]
- 23.Elgin S.C. and Workman, J.L. 2002. Chromosome and expression mechanisms: A year dominated by histone modifications, transitory and remembered. Curr. Opin. Genet. Dev. 12: 127-129. [DOI] [PubMed] [Google Scholar]
- 24.FlyBase. 2002. www.flybase.bio.indiana.edu.
- 25.Gatti M. and Pimpinelli, S. 1992. Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 26: 239-275. [DOI] [PubMed] [Google Scholar]
- 26.Grady D.L., Ratliff, R.L., Robinson, D.L., McCanlies, E.C., Meyne, J., and Moyzis, R.K. 1992. Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl. Acad. Sci. 89: 1695-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haaf T. and Willard, H.F. 1998. Orangutan α-satellite monomers are closely related to the human consensus sequence. Mamm. Genome 9: 440-447. [DOI] [PubMed] [Google Scholar]
- 28.Hari K.L., Cook, K.R., and Karpen, G.H. 2001. The Drosophila Su(var)2–10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15: 1334-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haupt W., Fischer, T.C., Winderl, S., Fransz, P., and Torres-Ruiz, R.A. 2001. The centromere1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27: 285-296. [DOI] [PubMed] [Google Scholar]
- 30.Hendrich B.D. and Willard, H.F. 1995. Epigenetic regulation of gene expression: The effect of altered chromatin structure from yeast to mammals. Hum. Mol. Genet. 4 Spec No: 1765-1777. [DOI] [PubMed] [Google Scholar]
- 31.Hochstenbach R., Harhangi, H., Schouren, K., and Hennig, W. 1994. Degenerating gypsy retrotransposons in a male fertility gene on the Y chromosome of Drosophila hydei. J. Mol. Evol. 39: 452-465. [DOI] [PubMed] [Google Scholar]
- 32.Horvath J.E., Schwartz, S., and Eichler, E.E. 2000a. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvath J.E., Viggiano, L., Loftus, B.J., Adams, M.D., Archidiacono, N., Rocchi, M., and Eichler, E.E. 2000b. Molecular structure and evolution of an α satellite/non-α satellite junction at 16p11. Hum. Mol. Genet. 9: 113-123. [DOI] [PubMed] [Google Scholar]
- 34.Howe M., Dimitri, P., Berloco, M., and Wakimoto, B.T. 1995. Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster. Genetics 140: 1033-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jabs E.W. and Persico, M.G. 1987. Characterization of human centromeric regions of specific chromosomes by means of alphoid DNA sequences. Am. J. Hum. Genet. 41: 374-390. [PMC free article] [PubMed] [Google Scholar]
- 36.Jabs E.W., Goble, C.A., and Cutting, G.R. 1989. Macromolecular organization of human centromeric regions reveals high-frequency, polymorphic macro DNA repeats. Proc. Natl. Acad. Sci. 86: 202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenuwein T. and Allis, C.D. 2001. Translating the histone code. Science 293: 1074-1080. [DOI] [PubMed] [Google Scholar]
- 38.John B. 1988. The biology of heterochromatin. In Heterochromatin: Molecular and structural aspects (ed. R.S. Verma), pp. 1–147. Cambridge University Press, Cambridge.
- 39.Karpen G.H. and Allshire, R.C. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489-496. [DOI] [PubMed] [Google Scholar]
- 40.Karpen G.H. and Spradling, A.C. 1990. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell 63: 97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karpen G.H. and Spradling, A.C. 1992. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132: 737-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karpen G.H., Le, M.H., and Le, H. 1996. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis [see comments]. Science 273: 118-122. [DOI] [PubMed] [Google Scholar]
- 43.Koch J. 2000. Neocentromeres and α satellite: A proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 9: 149-154. [DOI] [PubMed] [Google Scholar]
- 44.Kotani H., Hosouchi, T., and Tsuruoka, H. 1999. Structural analysis and complete physical map of Arabidopsis thaliana chromosome 5 including centromeric and telomeric regions. DNA Res. 6: 381-386. [DOI] [PubMed] [Google Scholar]
- 45.Kumekawa N., Hosouchi, T., Tsuruoka, H., and Kotani, H. 2000. The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7: 315-321. [DOI] [PubMed] [Google Scholar]
- 46.Lander E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 47.Le M.H., Duricka, D., and Karpen, G.H. 1995. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics 141: 283-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee C., Critcher, R., Zhang, J.G., Mills, W., and Farr, C.J. 2000. Distribution of γ satellite DNA on the human X and Y chromosomes suggests that it is not required for mitotic centromere function. Chromosoma 109: 381-389. [DOI] [PubMed] [Google Scholar]
- 49.Lo A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H. 2001a. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. Embo. J. 20: 2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo A.W.I., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H.A. 2001b. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11: 448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohe A.R. and Brutlag, D.L. 1986. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc. Natl. Acad. Sci. 83: 696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohe A.R., Hilliker, A.J., and Roberts, P.A. 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134: 1149-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Losada A., Abad, J.P., and Villasante, A. 1997. Organization of DNA sequences near the centromere of the Drosophila melanogaster Y chromosome. Chromosoma 106: 503-512. [DOI] [PubMed] [Google Scholar]
- 54.Maggert K.A. and Karpen, G.H. 2001. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158: 1615-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKee B.D. and Karpen, G.H. 1990. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell 61: 61-72. [DOI] [PubMed] [Google Scholar]
- 56.Moore D.P. and Orr-Weaver, T.L. 1998. Chromosome segregation during meiosis: Building an unambivalent bivalent. Curr. Top Dev. Biol. 37: 263-299. [DOI] [PubMed] [Google Scholar]
- 57.Mravinac B., Plohl, M., Mestrovic, N., and Ugarkovic, D. 2002. Sequence of PRAT satellite DNA “frozen” in some Coleopteran species. J. Mol. Evol. 54: 774-783. [DOI] [PubMed] [Google Scholar]
- 58.Mullenbach R., Pusch, C., Holzmann, K., Suijkerbuijk, R., and Blin, N. 1996. Distribution and linkage of repetitive clusters from the heterochromatic region of human chromosome 22. Chromosome Res. 4: 282-287. [DOI] [PubMed] [Google Scholar]
- 59.Murphy T.D. and Karpen, G.H. 1995a. Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell 81: 139-148. [DOI] [PubMed] [Google Scholar]
- 60.Murphy T.D. and Karpen, G.H. 1995b. Localization of centromere function in a Drosophila minichromosome. Cell 82: 599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy T.D. and Karpen, G.H. 1998. Centromeres take flight: α satellite and the quest for the human centromere. Cell 93: 317-320. [DOI] [PubMed] [Google Scholar]
- 62.Myers E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H., Remington, K.A., et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196-2204. [DOI] [PubMed] [Google Scholar]
- 63.Pimpinelli S., Berloco, M., Fanti, L., Dimitri, P., Bonaccorsi, S., Marchetti, E., Caizzi, R., Caggese, C., and Gatti, M. 1995. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. 92: 3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roseman R.R., Johnson, E.A., Rodesch, C.K., Bjerke, M., Nagoshi, R.N., and Geyer, P.K. 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.SanMiguel P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43-45. [DOI] [PubMed] [Google Scholar]
- 66.Satinover D.L., Vance, G.H., Van Dyke, D.L., and Schwartz, S. 2001. Cytogenetic analysis and construction of a BAC contig across a common neocentromeric region from 9p. Chromosoma 110: 275-283. [DOI] [PubMed] [Google Scholar]
- 67.Schueler M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109-115. [DOI] [PubMed] [Google Scholar]
- 68.Sullivan B.A., Blower, M.D., and Karpen, G.H. 2001. Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2: 584-596. [DOI] [PubMed] [Google Scholar]
- 69.Sullivan K.F. 2001. A solid foundation: Functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11: 182-188. [DOI] [PubMed] [Google Scholar]
- 70.Sun X., Wahlstrom, J., and Karpen, G. 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi K., Murakami, S., Chikashige, Y., Funabiki, H., Niwa, O., and Yanagida, M. 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3: 819-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson-Stewart D., Karpen, G.H., and Spradling, A.C. 1994. A transposable element can drive the concerted evolution of tandemly repetitious DNA. Proc. Natl. Acad. Sci. 91: 9042-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapitz P., Glatzer, K.H., and Bunemann, H. 1992. Towards a physical map of the fertility genes on the heterochromatic Y chromosome of Drosophila hydei: Families of repetitive sequences transcribed on the lampbrush loops nooses and threads are organized in extended clusters of several hundred kilobases. Mol. Gen. Genet. 235: 221-234. [DOI] [PubMed] [Google Scholar]
- 74.Vafa O. and Sullivan, K.F. 1997. Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7: 897-900. [DOI] [PubMed] [Google Scholar]
- 75.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 76.Vig B.K. 1994. Do specific nucleotide bases constitute the centromere? Mutat. Res. 309: 1-10. [DOI] [PubMed] [Google Scholar]
- 77.Wakimoto B.T. 1998. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell 93: 321-324. [DOI] [PubMed] [Google Scholar]
- 78.Wallrath L.L. and Elgin, S.C. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263-1277. [DOI] [PubMed] [Google Scholar]
- 79.Warburton P.E., Haaf, T., Gosden, J., Lawson, D., and Willard, H.F. 1996. Characterization of a chromosome-specific chimpanzee α satellite subset: Evolutionary relationship to subsets on human chromosomes. Genomics 33: 220-228. [DOI] [PubMed] [Google Scholar]
- 80.Weiler K. and Wakimoto, B. 1996. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577-605. [DOI] [PubMed] [Google Scholar]
- 81.Willard H.F. 1990. Centromeres of mammalian chromosomes. Trends Genet. 6: 410-416. [DOI] [PubMed] [Google Scholar]
- 82.Willard H.F., Wayne, J.S., Skolnick, M.H., Schwartz, C.E., Powers, V.E., and England, S.B. 1986. Detection of restriction fragment length polymorphisms at the centromeres of human chromosomes by using chromosome-specific α satellite DNA probes: Implications for development of centromere-based genetic linkage maps. Proc. Natl. Acad. Sci. 83: 5611-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan C.M., Dobie, K.W., Le, H.D., Konev, A.Y., and Karpen, G.H. 2002. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics 161: 217-229. [DOI] [PMC free article] [PubMed] [Google Scholar]