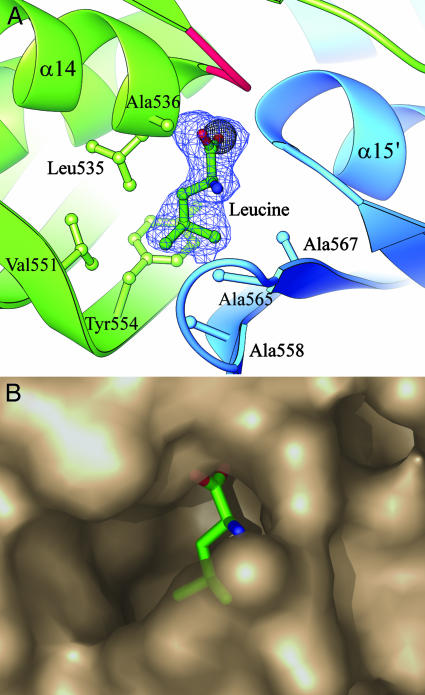
Fig. 3.
Leucine binding site for feedback inhibition. (A) Leucine is shown built into its electron density (2Fo - Fc omit map, contoured at 1σ) in the leucine complex structure. The carboxyl group displaces a chloride ion (gray sphere) seen in the leucine-free substrate structure, between the N-termini of helices α14 and α15′. Mutation of residues equivalent to Gly-531, Gly-533 (in the loop colored red), and Ala-536 abolish feedback inhibition in S. cerevisiae α-IPMS. Also shown are the residues that form the hydrophobic cavity for the isopropyl side chain. (B) Semitransparent PYMOL (DeLano Scientific, www.pymol.org) surface diagram of the leucine binding pocket.