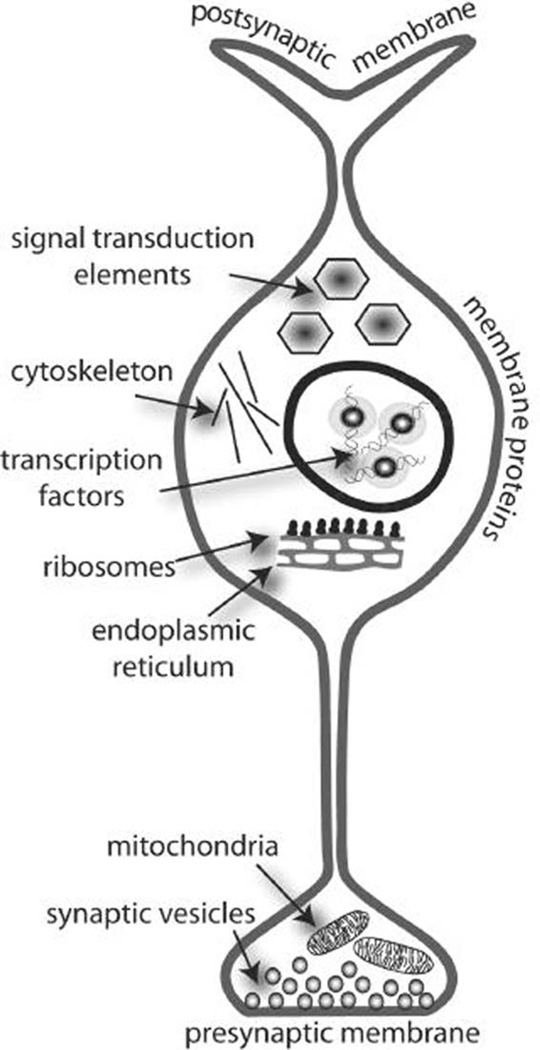
Fig. 3.
To put gene array or proteomics data into biological contexts, genes or proteins can be grouped according to their cellular location (i.e., membrane, postsynaptic membrane, mitochondrion, etc.) or their function (i.e., transcription factor, signal transduction element, mitochondrial respiration, vesicular fusion, etc.). Furthermore, functions can overlap with cellular locations, such as ‘mitochondrion’ and ‘mitochondrial respiration’, ‘nucleus’ and ‘transcription factors’. Computer programs are available that calculate hypergeometric distributions to determine if particular functions or locations are more affected than would be expected by chance.