Abstract
Determining the molecular mechanism(s) leading to Purkinje neuron loss in the neurodegenerative disorder fragile X-associated tremor/ataxia syndrome (FXTAS) is limited by the complex morphology of this cell type. Purkinje neurons are notoriously difficult to isolate and maintain in culture presenting considerable difficultly to identify molecular changes in response to expanded CGG repeat (rCGG)-containing mRNA that induces neurotoxicity in FXTAS. Several studies have uncovered a number of RNA-binding proteins involved in translation that aberrantly interact with the CGG-containing RNA; however, whether these interactions alter the translational profile of cells has not been investigated. Here we employ bacTRAP translational profiling to demonstrate that Purkinje neurons ectopically expressing 90 CGG repeats exhibit a dramatic change in their translational profile even prior to the onset of rCGG-induced phenotypes. This approach identified ∼500 transcripts that are differentially associated with ribosomes in r(CGG)90-expressing mice. Functional annotation cluster analysis revealed broad ontologies enriched in the r(CGG)90 list, including RNA binding and response to stress. Intriguingly, a transcript for the Tardbp gene, implicated in a number of other neurodegenerative disorders, exhibits altered association with ribosomes in the presence of r(CGG)90 repeats. We therefore tested and showed that reduced association of Tardbp mRNA with the ribosomes results in a loss of TDP-43 protein expression in r(CGG)90-expressing Purkinje neurons. Furthermore, we showed that TDP-43 could modulate the rCGG repeat-mediated toxicity in a Drosophila model that we developed previously. These findings together suggest that translational dysregulation may be an underlying mechanism of rCGG-induced neurotoxicity in FXTAS.
INTRODUCTION
Fragile X-associated tremor/ataxia syndrome (FXTAS) is an inherited neurodegenerative disorder characterized by progressive intention tremor, gait ataxia and cognitive decline. FXTAS predominantly affects older male carriers of premutation expansion alleles (55–200 CGG repeats) in the 5′ untranslated region (UTR) of the fragile X mental retardation 1 (FMR1) gene (1). FXTAS penetrance is largely dependent on carrier age, with symptoms typically seen in 30% of males by the age of 50 and in 50% of males older than 70 years (2). Individuals with FXTAS develop a common radiological profile that includes global brain atrophy and white-matter disease (3). The neuropathological hallmarks of FXTAS observed upon autopsy include the presence of large eosinophilic, ubiquitin-positive intranuclear inclusions found in neurons and astrocytes throughout the brain along with axonal degeneration of cerebellar Purkinje neurons (4,5).
Premutation alleles are transcribed to produce mRNAs carrying expanded CGG repeats (rCGG) that can be found in RNA foci (6) and/or inclusions (7). These mRNAs are hypothesized to initiate damage to neurons in FXTAS, but the precise mechanism(s) are not well understood. Studies of the toxic gain of RNA function in the myotonic dystrophies (DM1 and DM2) have determined that the expanded CUG or CCUG repeats assume structures that bind, sequester and functionally deplete the free pool members of the MBNL protein family in affected cells. MBNL protein depletion leads to misregulated alternative splicing of its mRNA targets leading to a number of DM phenotypes. In addition, alterations to activities of members of the CELF family of RNA-binding proteins contribute to disease phenotypes (8,9). In the DMs, changes to RNA-binding protein functions lead to misregulation of transcription, splicing and translation of mRNAs, disrupting numerous cellular functions. In order to describe mechanism(s) by which rCGG repeats cause FXTAS, several studies have used genetic and biochemical approaches to identify RNA-binding proteins that interact with expanded rCGG repeats (10). Jin et al. developed a Drosophila model that expresses premutation-length r(CGG) repeats in fly photoreceptor cells. The resultant neurodegeneration can be suppressed by the overexpression of RNA-binding proteins CELF1, hnRNP A2/B1 or Purα (6,11–13). Nuclear accumulation of mRNAs involved in stress and immune response has been described (14), suggesting that impaired nuclear export could alter the translational profile of specific mRNAs. Sellier et al. demonstrated that RNA-binding protein Sam68 is present in FXTAS inclusions and subsequently loses its splicing regulatory function through depletion (6). These authors also uncovered >50 RNA-binding proteins, including MBNL1, present in RNA aggregates induced by rCGG expression in cultured cells. MicroRNA (miRNA)-processing components DROSHA and DGCR8 were also associated with aggregates, suggesting possible effects on miRNAs and their target mRNAs (15). For most of these proteins, their recruitment to aggregates was either transient or an end-stage event occurring just prior to cell death, suggesting that RNA-binding protein association may not be directly related to rCGG expression.
We previously generated a transgenic mouse model that directs the expression of 90 CGG repeats in cerebellar Purkinje neurons (16). In this model, Purkinje neurons develop ubiquitin-positive intranuclear inclusions, swollen axons with axonal torpedoes and begin to die by 4 months of age. These mice also develop an age-dependent, progressive decline in motor coordination assessed by accelerating rotarod. As with models in Drosophila (12), pathology is found whether the rCGG is expressed in the context of an Fmr1 cDNA or in an unrelated transcript, EGFP, demonstrating that the neuronal toxicity results from the expanded rCGG repeats independent of the remainder of the RNA transcript.
In both human FXTAS and our mouse model, Purkinje neurons die from the effects of rCGG expression. To begin to approach the mechanism of this toxicity, we sought to study the gene expression profile in young presymptomatic mice with no detectable ubiquitin-positive inclusions. Changes in the translational profiles of ribosome-associated mRNAs in 4-week-old rCGG mice were compared with age-matched control mice without CGG repeats. We utilized the translating ribosome affinity purification (TRAP) method (17) to immunoprecipitate (IP) actively translating mRNA transcripts specifically from Purkinje neurons and to obtain a snapshot of early translational changes that occur in response to CGG repeat expression. This approach identified ∼500 transcripts with altered ribosome association in response to rCGG expression in Purkinje neurons. Gene ontology (GO) analysis suggests that transcripts with increased association with ribosomes are enriched in biological processes such as synapse organization and cellular stress response whereas many of those mRNAs showing reduced association with ribosomes are involved in RNA processing. Among this latter set is the transcript for Tardbp, which encodes the protein TDP-43, an RNA-binding protein that is also a marker of neurodegeneration commonly found in inclusions in amyotrophic lateral sclerosis (ALS) (18). We find a significant loss in TDP-43 immunoreactivity in the Purkinje neurons of young rCGG-expressing mice and also demonstrate genetic interaction in the fly model. Expression of rCGG in Purkinje neurons leads to alterations in the translational profile of mRNAs in presymptomatic mice, supporting the hypothesis that rCGG repeats play a major role in FXTAS neuropathology even prior to the onset of symptoms or pathology.
RESULTS
Identifying translational changes in response to (CGG)90 expression
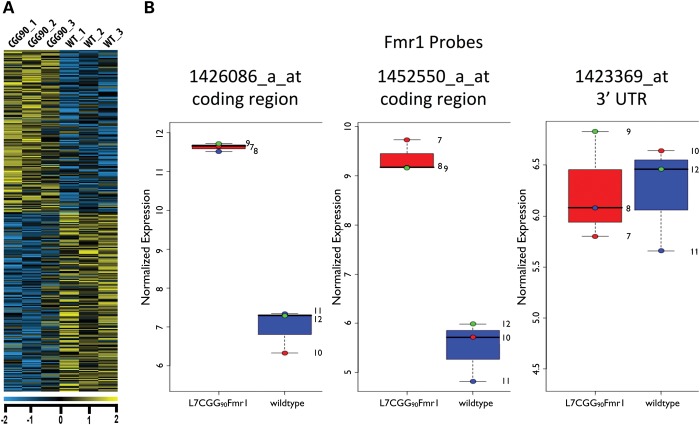
To identify transcripts with altered ribosome association in response to Fmr1 mRNA carrying an expanded (CGG)90 repeat, we performed microarray analysis on Purkinje neuron-specific ribosome-bound mRNAs isolated from mice expressing expanded repeats by the TRAP method (17), using hybridization to Affymetrix Mouse Genome 430 2.0 Arrays. In order to assess early translational changes and to reduce detection of potential secondary responses, presymptomatic 4-week-old mice expressing 90 CGG repeats in the 5′ UTR of a Purkinje neuron-specific transgene encoding Fmr1 [L7CGG90Fmr1 (16)] were compared with non-transgenic littermates. At 4 weeks, L7CGG90Fmr1 mice show normal Purkinje neuron morphology and exhibit no ubiquitin-positive intranuclear inclusions (data not shown). This finding is similar to the results with age-matched, wild-type (WT) littermate controls and control mice expressing Purkinje neuron-specific transgenes expressing Fmr1 without CGG repeats (16). We removed cerebella and isolated translating mRNAs specifically from Purkinje neurons for microarray analysis via TRAP. The microarray analysis revealed 498 transcripts with significantly altered polysome association in the presence of r(CGG)90 repeat mRNA (Fig. 1A). The transgene-expressed Fmr1 showed increased polysome association and acted as an internal positive control (Fig. 1B). The L7CGG90Fmr1 transgene is a hybrid of an Fmr1 coding cDNA with the bovine growth hormone 3′ UTR. As expected, the microarray probe that detects the Fmr1 3′ UTR was not significantly changed when comparing transgenic and non-transgenic animals, suggesting that the levels of the endogenous Fmr1 mRNA are unchanged in polysome abundance (Fig. 1B).
Figure 1.
Translational profile of r(CGG)90-expressing Purkinje neurons. (A) Heatmap showing cerebellar translational profiles from Purkinje neurons of L7CGG90Fmr1 transgenic and WT control mice [littermates without the r(CGG)90 transgene], respectively. Each column represents a Purkinje neuron-specific IP pool from the cerebella of four mice. Each row represents one gene. Expression levels are depicted according to the color scale at the bottom (Bayes-corrected P < 0.05). (B) The graphs show the results of three sets of experiments (each set consisting of an immunoprecipitation from four pooled cerebella of WT control which express L7EGFPL10a and four pooled cerebella of experimental mice which express L7CGG90Fmr1 and L7EGFPL10a), paired to include a WT and experimental immunoprecipiation performed on the same date and denoted by red, blue and green color-coded dots. The GCRMA normalized log2 values for each of three individual Fmr1 probes present on the Mouse 430 2.0 microarray reveal that the internal positive control, Fmr1, exhibits a significantly elevated association with ribosomes over WT. The probe set representing the 3′ UTR usage of Fmr1 shows that endogenous levels of Fmr1 are unchanged between WT and L7CGG90Fmr1 expressing Purkinje neurons.
Analysis of mRNAs exhibiting altered polysome abundance in Purkinje neurons expressing CGG90 RNA
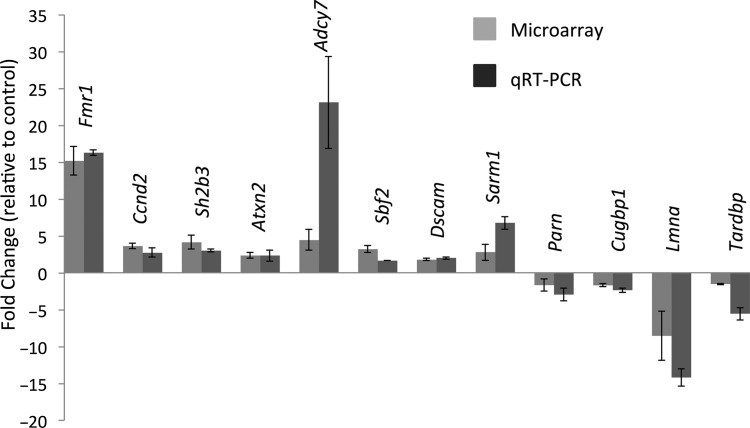
Of those transcripts with significantly altered polysome association (P < 0.05), 261 transcripts showed increased polysome abundance whereas 237 were decreased in Purkinje neurons from the (CGG)90 model (Supplementary Material, Table S1). To validate these results, we measured the levels of 14 mRNAs in Purkinje neuron ribosome preparations by qRT-PCR, comparing the CGG90 mice with littermates lacking the CGG90 transgene. We confirmed alterations (P < 0.05) in 12 of 14 mRNAs tested (86%) (Fig. 2). The two mRNA species that did not meet statistical significance in the qRT-PCR analysis nonetheless showed a trend consistent with the microarray analysis results (data not shown). As expected, Fmr1 is found at up to 15-fold higher levels on the Purkinje neuron ribosomes of the (CGG)90 expressing mice compared with WT Purkinje neurons.
Figure 2.
Validation of microarray hits by qRT-PCR on Purkinje neuron RNA. Fourteen genes from the microarray experiment, from a range of expression level changes, were tested by real-time PCR to validate the expression changes. All genes showed an expression change in the same direction as the microarray. All genes are significant to 95% confidence when tested by the student's t-test at the P < 0.05 level.
Biological functions of transcripts with (CGG)90 induced alterations in ribosome association
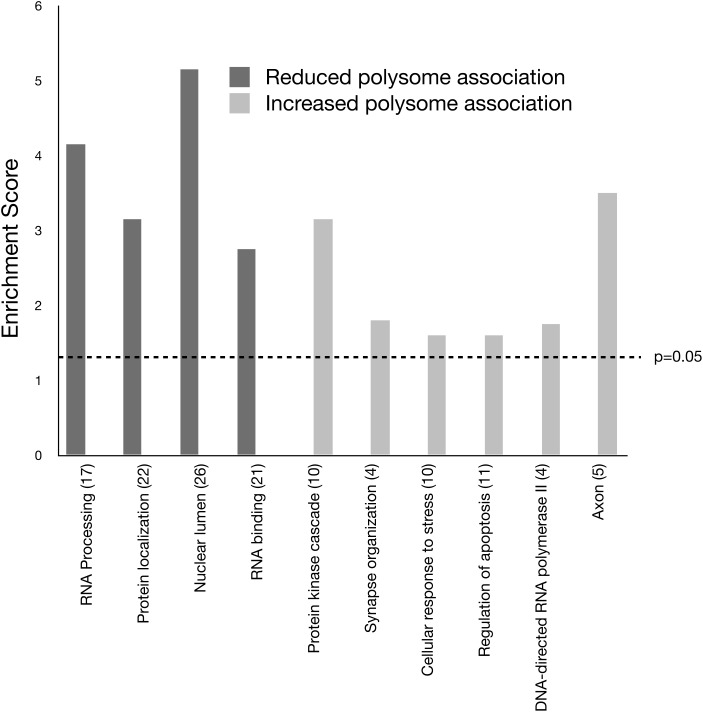
We performed a GO analysis using the Database for Annotation, Visualization and Integrated Discovery to obtain insight into the biological functions of mRNAs with altered ribosome association in response to (CGG)90 expression (19,20). For this analysis, we separated the data into two groups: transcripts exhibiting elevated ribosome association in response to (CGG)90 repeat expression and transcripts exhibiting reduced ribosome association in response to (CGG)90 repeat expression. We utilized GO_FAT to identify statistically enriched functional groups. Several broad GO terms were enriched in response to (CGG)90 expression including RNA binding, protein localization and protein kinase cascade (Fig. 3). RNA-binding proteins were particularly interesting targets for potential mechanisms describing early events leading to rCGG-induced toxicity because they could implicate mRNA targets with misregulated RNA processing. The transcripts of 22 RNA-binding protein genes were reduced on polysomes in CGG90 Purkinje neurons. Among these are Celf1, which is implicated in DM pathology and found to modify the neurodegenerative eye phenotype in the fly models of FXTAS (13), and Tardbp, encoding TDP-43, an RNA-binding protein that has been found mutated in neurodegenerative disorders ALS and frontotemporal degeneration (ALS/FTD) (18). TDP-43 is also a prominent protein component of neuronal inclusions in these disorders. Increased polysome association of mRNAs involved in cellular response to stress suggests that Purkinje neurons may already be responding to stress resulting from the expanded r(CGG) repeat expression by 4 weeks of age.
Figure 3.
Analysis of GO categories for genes whose mRNAs are differentially associated with ribosomes in the Purkinje neurons of the r(CGG)90 mouse model. The bar graph shows the GO terms and the number of genes in each category that are significantly enriched in the r(CGG)90 gene list. The enrichment score (E) is the P-value in the −log scale. The P-value shows whether the gene list is specifically enriched in the corresponding annotation cluster compared with random chance. The enrichment score is ≥1.3 (dashed line) when the P-value is ≤0.05.
TDP-43 abundance is reduced in (CGG)90 Purkinje neurons
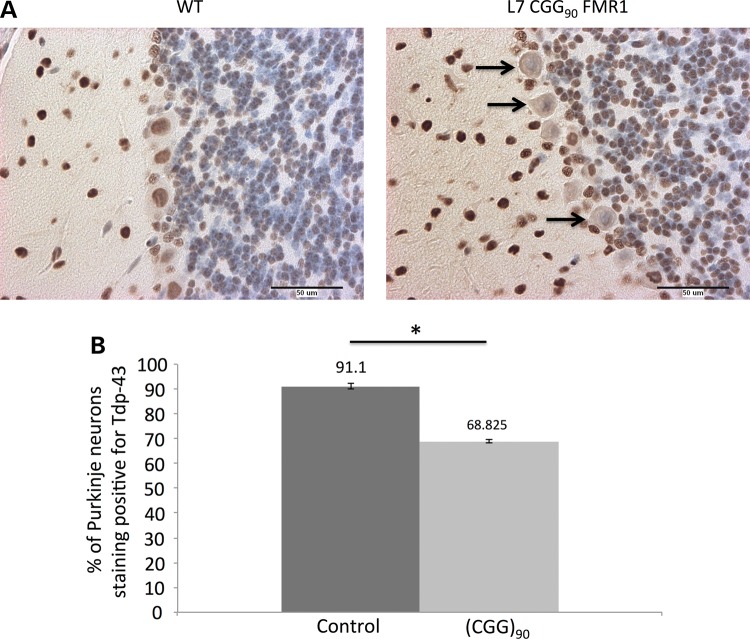
To determine whether rCGG-induced changes in the translational profile also led to an altered protein expression profile, we investigated the expression of previously validated targets (see Supplementary Material, Table S2) in Purkinje neurons using immunostaining. Of the proteins we tested, TDP-43 was the only one to exhibit a detectable change in protein expression. Our microarray analysis indicated a significantly reduced level of ribosome-associated Tardbp mRNA in Purkinje neurons of mice expressing 90 CGG repeats. The Tardbp gene product, TDP-43, is found in neuronal protein aggregates in FTD and ALS. To determine whether this change in polysome association led to reduced levels of TDP-43 protein, we investigated levels and/or localization of TDP-43 protein in Purkinje neurons using immunostaining. Sagittal sections of brains isolated from mice expressing (CGG)90 RNA were incubated with an antibody that detects TDP-43. In animals at 4 weeks of age, we saw a significant reduction in the number of Purkinje neuron cell bodies staining positive for TDP-43. This result fits the predicted direction from the microarray and mRNA data but took the form of an increase in the number of cells with undetectable protein, rather than an overall reduction in protein levels (Fig. 4A). Quantification of TDP-43-positive Purkinje neurons showed that the number of TDP-43-positive neurons was significantly decreased (P < 0.05) in (CGG)90-expressing neurons compared with control animals (Fig. 4B) at 4 weeks. We did not find a significant difference in TDP-43 immunoreactivity or total number of Purkinje neurons in 8-week-old (CGG)90-expressing Purkinje neurons compared with control Purkinje neurons (Supplementary Material, Fig. S1). Thus, the microarray analysis revealed an apparently transient decrease in TDP-43 in 4-week-old presymptomatic CGG90 Purkinje neurons as measured by immunohistochemistry.
Figure 4.
Loss of TDP-43 immunoreactivity in r(CGG)90-expressing Purkinje neurons. (A) Sagittal cerebellar sections of WT control and L7CGG90Fmr1 Purkinje neurons stained with antibody against TDP-43. Arrows denote Purkinje neurons in L7CGG90Fmr1 mice that have lost TDP-43 immunoreactivity (20× magnification). Size bar equals 50 µm. (B) The bar graph shows the percentage of Purkinje neurons that are immunoreactive for TDP-43 in sagittal cerebellar sections from a total of four r(CGG)90-expressing mice compared with four WT controls. Error bars indicate SEM. The symbol ‘*’ indicates that comparing positively staining Purkinje neurons between r(CGG)90 and WT using a t-test results in a P value of <0.0001.
TDP-43 modulates the rCGG repeat-mediated neurodegeneration in a Drosophila model of FXTAS
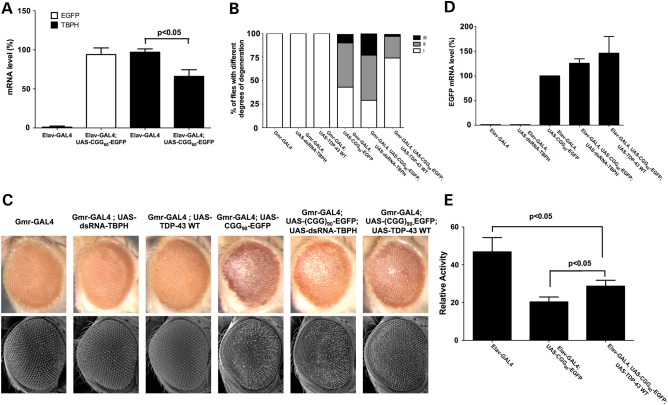
To further determine the functional significance of this reduced TDP-43 expression in rCGG repeat-mediated neurodegeneration, we utilized the Drosophila model that we had previously developed for the further functional analyses. Quantitative RT-PCR analyses revealed that the expression of rCGG repeat in fruit fly could also down-regulate the expression level of endogenous TDP-43 fly ortholog, TBPH (Fig. 5A). We have previously shown that premutation-length rCGG repeats can cause the neurodegenerative changes to fly photoreceptors (12). To test the genetic interaction between rCGG repeat-mediated toxicity and TDP-43, we crossed Gmr-GAL4, UAS-(CGG)90-EGFP transgenic flies with flies carrying both a UAS-TDP-43 transgene and a UAS-dsRNA-TBPH construct capable of overexpression or knockdown of expression of TDP-43 or its endogenous ortholog. We found that the expression of WT TDP-43 could suppress neurodegeneration, whereas knockdown of the endogenous TDP-43 fly ortholog, TBPH, enhanced the eye phenotype (Fig. 5B and C). Expression of TDP-43 or knockdown of TBPH had no effect on the expression of the rCGG repeat (Fig. 5D). Additionally, using the locomotion assay (21), we observed similar, albeit smaller, suppression of rCGG repeat toxicity mediated by the expression of WT TDP-43. These results together suggest that the reduced expression of TDP-43 that we observed in FXTAS transgenic mice plays a significant role in rCGG repeat-mediated neurodegeneration.
Figure 5.
TDP-43 could modulate rCGG repeat-mediated neurodegeneration in Drosophila. (A) Quantitative RT-PCR was used to determine the expression levels of EGFP and TBPH in fly heads expressing rCGG60 repeats. The relative expression levels as determined by ΔΔCt analyses are shown. Values are mean ± SD for triplicate samples. (B and C) Expression of the rCGG90 repeats disrupts the Drosophila eye morphology by light (B) and scanning electron microscope (C). All flies shown are 2 weeks old after eclosion. Quantification of Drosophila eye disruption after 2 weeks is shown in C. Eye defects are grouped into three categories: I: <25% ommatidia loss, II: 25–50% ommatidia loss and III: >50% ommatidia loss with necrosis. (D) Quantitative RT-PCR was used to determine the expression level of EGFP in fly heads expressing rCGG60 repeats. The relative expression levels as determined by ΔΔCt analyses are shown. Values are mean ± SD for triplicate samples. (E) Flies were exposed to standard 12 : 12 light/dark cycle at 25°C in DAM system, and their activity is measured. At least 70 flies per genotype were pooled to obtain the group averages and error bars (standard error of the mean) shown.
DISCUSSION
In the present study, we tested whether Purkinje neuron translational profiles are altered in response to r(CGG)90 repeat expression. Because the mechanism(s) of RNA toxicity contributing to FXTAS and the alterations to neuronal metabolism that result from the presence of expanded r(CGG) repeats remain unclear, our data provide insights into the nature of the in vivo response of this critical cerebellar neuron as defined by its translational profile. We identified 498 transcripts with altered polysome association in young Purkinje neurons expressing an expanded r(CGG)90 repeat mRNA. These included RNA-binding proteins and RNA processing genes. To our knowledge, these data provide the first evidence that expanded CGG repeat expression leads to alterations in translational profiles prior to the onset of neuropathology, which could be an early event in the development of FXTAS.
Intriguing recent evidence suggests that premutation carriers can exhibit symptoms prior to the onset of clinical features of FXTAS. Premutation carriers exhibit executive dysfunction and anxiety that increases with age prior to FXTAS onset (22,23). Reports of associated memory deficit (24), ADHD (25), psychopathological disturbances (24,26) and autism (27–29) in premutation carriers without FXTAS suggests that there may also be developmental effects resulting from FMR1 premutations. In support of the idea of a pre-FXTAS neurodevelopmental phenotype, a recent study by Cunningham et al. demonstrated that knock-in mouse embryos (carrying a CGG repeat expansion in their Fmr1 gene) exhibit neocortical migration defects and altered expression of neuronal lineage markers (30), suggesting that the premutation allele can lead to alterations in brain development. Chen et al. showed that hippocampal neurons cultured from young knock-in mice exhibit early developmental defects such as shortened dendrites and alterations in synaptic morphology (31). In addition, Diep and colleagues found that young female knock-in mice exhibit a decline in motor coordination that correlates with increasing CGG repeat length (32).
The translational profile of the Purkinje neuron-specific mouse model revealed changes in mRNAs for genes implicated in FXTAS pathology. For example, Lamin A/C is present in both the inclusions of FXTAS patients and the inclusions in cell culture (33,34). Interestingly, mRNA from FXTAS patient frontal cortex revealed a 2-fold elevation in LMNA mRNA levels over control individuals without the premutation whereas LMNA protein expression was reduced to undetectable levels in RIPA-soluble protein extracts from the CNS panel of eight out of ten FXTAS patients (35). Reduction in soluble LMNA appeared to be a consequence of repartitioning of less soluble or possibly aggregated forms of LMNA that might contribute to inclusion formation. Our results demonstrate that Lmna mRNA has reduced association with ribosomes in Purkinje neurons in response to r(CGG)90. It would be interesting to determine whether a potential mechanism for LMNA reduction in FXTAS is also linked to reduced ribosome association and/or translational efficiency of LMNA transcripts.
Prior data suggest a role for CELF1 in the pathogenesis of FXTAS. In myotonic muscular dystrophy, CELF1 steady state levels are increased. Increasing the level of CELF1 in the Drosophila model of FXTAS suppresses the r(CGG)90-mediated neurodegenerative eye phenotype (13). Our data predict that CELF1 protein expression is reduced in the presence of r(CGG)90. In addition to its splicing regulatory functions, CELF1 has been implicated in several other RNA metabolic processes including mRNA degradation. In muscle cells, depletion of CELF1 or expression of expanded DMPK 3′ UTR transcripts is sufficient to stabilize specific mRNA transcripts (36). CELF1 binds 3′ UTR elements of its RNA substrates and recruits the deadenylase, Parn, in vitro (37) to destabilize mRNAs. Our data predict that Parn protein expression is also reduced in response to r(CGG)90 expression. A decrease in translation of Celf1 and/or Parn mRNA presents the potential for a new mechanism for the pathogenesis of FXTAS where altered splicing of CELF1 targets or stabilization of mRNAs owing to r(CGG)90 expression leads to symptoms. It would be interesting to compare the list of mRNAs we identified that show reduced ribosome association with that of neuron- or brain-specific CELF1 mRNA targets.
Mutations in TARDBP have been linked to the neurodegenerative disorders ALS and FTD, now known as FTLD-TDP (18). This RNA/DNA-binding protein is a major component of the neuronal aggregates in the familiar forms of the disorders (38); however, TDP-43-positive inclusions have also been reported in cases of Alzheimer disease, Pick disease, dementia with Lewy bodies and other neurodegenerative disorders and are seen in glia as well as neurons (18). The mechanisms by which TDP-43 mutations cause neurodegeneration remain under investigation. In healthy cells, TDP-43 is found primarily in the nucleus, but patients with TDP-43 proteinopathy as in ALS and FTD with ubiquitin-positive inclusions exhibit a redistribution of TDP-43 from the nucleus to the cytoplasm (38). The cytoplasmic redistribution of TDP-43 is proposed to be an early event in the formation of TDP-43-positive inclusions in addition to the appearance of ‘pre-inclusions’ or granular cytoplasmic TDP-43 aggregates (39–42). Additionally, neurons with pre-inclusions lose nuclear TDP-43 immunoreactivity (43). We did not see a redistribution of TDP-43 protein to the cytoplasm or the formation of cytoplasmic ‘pre-inclusions’ in young r(CGG)90 mice. Instead, we found a significantly reduced fraction of r(CGG)90-expressing Purkinje neurons with detectable TDP-43. Recent data demonstrate that axonal transport of TDP-43-containing mRNA/protein particles is impaired by ALS-causing mutations (44). This observation suggests a mechanism for alterations in TDP-43 abundance leading to neurodegeneration through altered localization mRNA targets.
Of the proteins we assayed for altered immunoreactivity in response to r(CGG)90 RNA, TDP-43 was the only one to show altered ribosome association together with detectably altered expression at the level of immunohistochemistry. We found a greater than a 6-fold decrease in Tardbp mRNA associated with ribosomes in 4-week-old mice expressing r(CGG)90 in Purkinje neurons. This transcript is one of the most down-regulated that we observed. Intriguingly, we did not find a significant difference in TDP-43 immunoreactivity in 8-week-old r(CGG)90 mice compared with WT animals. A possible explanation could be that those neurons that lose TDP-43 immunoreactivity in 4-week-old mice could have died and were, therefore, not counted. However, we could not detect a statistically significant difference in the total number of Purkinje neurons between 4- and 8-week-old WT control and r(CGG)90 mice nor did we find a statistically significant difference in the totally number of Purkinje neurons between WT control or r(CGG)90 within the respective age groups. The loss and recovery of TDP-43 in r(CGG)90-expressing neurons may represent a delay in the developmental onset of TDP-43 expression; however, the fact that the loss of TDP-43 immunoreactivity is in agreement with the microarray results suggests that Tardbp transcripts exhibit an early reduction in translation in response to r(CGG)90.
The functional relevance of reduced expression of TDP-43 in FXTAS pathogenesis was further supported by our genetic interaction experiment in Drosophila. Using different assays, we show that the co-expression of WT TDP-43 can suppress the rCGG repeat-mediated neurodegeneration, indicating the TDP-43-mediated gene regulation could be altered by fragile X premutation rCGG repeats. It would be interesting to further test this using a TDP-43 transgenic mouse model. Given the role of TDP-43 in RNA metabolism, it would be important to identify the RNAs regulated by TDP-43 that could contribute to the molecular pathogenesis of FXTAS.
It will also be interesting to determine whether there are alterations in TDP-43 levels in lymphoblasts or fibroblasts of presymptomatic and symptomatic premutation carriers. While presymptomatic brain tissue from premutation carriers is likely unavailable or limited, studies of TDP-43 levels at autopsy of older premutation carriers could help us to better understand how these results are related to TDP-43 expression in patients. However, it is important to note the caveat that these studies may only be relevant to brain tissue and at early ages.
In summary, several functional classes of genes were differentially translated in Purkinje neurons ectopically expressing 90 rCGG repeats. These ontology classes provide insight into molecular pathways that respond to the toxic CGG-repeat RNA. For example, transcripts having decreased association with ribosomes fall into functional categories such as RNA processing, RNA splicing and protein localization. Loss of regulation of these processes has been previously implicated in a host of repeat expansion disorders such as myotonic dystrophy, spinocerebellar ataxias 3, 8 and 10 and Huntington disease-like 2 and ALS/FTD (9,45). The list of transcripts we found with altered ribosome association can potentially be used to define molecular biomarkers of presymptomatic states of FXTAS, present several new candidate genes for the study of genetic modifiers, provide a list of potential therapeutic targets for the treatment of FXTAS and might reveal common pathways between other Purkinje neuron degeneration diseases.
MATERIALS AND METHODS
Mouse husbandry
L7CGG90Fmr1:Pcp2 BAC (r(CGG)90 BAC) and wild-type:L7/Pcp2 BAC (WT BAC) transgenic mice used in this study have a 50% C57B6 and 50% FVB genetic background and were derived from the same parents by crossing L7CGG90Fmr1 heterozygous transgenics (100% C57B6 background) with L7/Pcp2eEGFPL10a BAC homozygous transgenics (100% FVB background). All mice reported were F1 progeny and were therefore hemizygous for one or both transgenes. Littermate WT mice were used for all experiments whenever possible. All experiments were conducted in accordance with the NIH Guide for the Use and Care of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Immunoprecipitation, quantification and amplification of mRNA samples
Purification of translating mRNAs from Purkinje neurons was performed as previously described (17) using a mix of two monoclonal antibodies (19C8 and 19F7). Four mice, two male and two female, for each replicated sample were euthanized with CO2, and their cerebella were dissected. Each genotype was assayed in triplicate. Purified mRNA samples were analyzed using a Nanodrop 1000 spectrophotometer and Agilent 2100 Bioanalyzer (Foster City, CA, USA) in order to assess mRNA quality and quantity as reflected by rRNA levels and integrity. For each sample, the purified RNA was converted to double-stranded cDNA using the NuGEN Technologies (San Carlos, CA, USA) Ovation Pico WTA kit for nucleic acid amplification followed by fractionation and biotin labeling using the NuGEN Encore Biotin Module prior to hybridization to Affymetrix 430 2.0 microarrays according to the manufacturer's instructions.
Microarray normalization and data analysis
Low-level Affymetrix Cel files were normalized using the GCRMA method, which produces probe-set summaries on the log2 scale after accounting for oligo-level intensity variation and accounting for GC content of probes (46). Since most probes on the array are not expressed in the specific cell type and are not captured in the IP step, we expect most probe-level summaries to correspond to nonspecific background. We used a kernel density estimate visualization of the probe-set means across the arrays (GSE57034) to determine an appropriate cut-off; based on the inflection point visible in the density plot, we identified a cut-off that corresponds to an overall mean value of at least 3.423 for the hybridizations considered in this study, corresponding to approximately the 60th percentile of the distribution. We determined 16 040 out of 45 101 probe-sets on the array that fulfilled this condition. Of these 16 040, 16 039 had an average above the 3.423 cut-off in the WT animals alone, and 16 037 had an average above the cut-off in the mutant alone. For the 16 040 probe-sets, we performed least-squares and a two-way ANOVA model to fit each probe set with IP date (three levels; one for each IP date) and Genotype (L7CGG90Fmr1 and WT/control) as explanatory factors. We then fit a linear contrast comparing the mean of (CGG)90 and WT, and we used the empirical Bayes method as implemented in the R package, LIMMA, to determine moderated T-statistics and P-values. We used a simple cut-off of marginal P-value of <0.05 to determine a set of 498 probe-sets for further analysis and validation. Because of the small size of our study, the false discovery rate adjustment using the Benjamini–Hochberg q-value level 0.05 was impractical; therefore, we relied on qPCR validation to determine the empirical false-positive rate of our findings.
Quantitative real-time RT-PCR
cDNA was synthesized from 15 ng of total mRNA from the three replicate IP samples with the NuGEN Ovation Pico WTA System and then purified with the QIAGEN Quick PCR cleanup, according to the manufacturer's instructions (QIAGEN, Valencia, CA, USA). TaqMan PrimeTime gene-specific predesigned assays (Integrated DNA Technologies, Coralville, IA, USA) were used following the manufacturer's instructions. Fluorescence was detected with the 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Each replicate was assayed in triplicate. Data were normalized to Gapdh using the comparative ddCT method.
Immunohistochemistry
Approximately four mice per age/genotype (wild type—4 weeks, wild type—8 weeks, CGG90—4 weeks and CGG90—8 weeks) were sacrificed by cervical dislocation; brains were dissected immediately and fixed overnight in 10% neutral buffered formalin for 24 h followed by paraffin embedding according to standard protocols. Three sagittal sections (6 µm) per brain were deparaffinized, followed by antigen retrieval using microwave treatment in 0.01 m sodium citrate solution. The sagittal sections were immunostained with the following antibodies and at the indicated dilutions: mouse anti-FMRP clone 1C3 (1:4000, Abcam), rabbit anti-ubiquitin (1:500, DAKO), rabbit anti-Lamin A/C (1:1000, GeneTex), rabbit anti-PARN (1:1000, Proteintech), rabbit anti-CUGBP1 (1:1000, Proteintech) and rabbit anti-TARDBP/TDP-43 (1:1000, Proteintech). Each Purkinje neuron of one section per animal was counted and assessed for changes in protein expression/localization compared with wild type. The percentage of Purkinje neurons showing changes in immunoreactivity in sagittal cerebellar sections from a combined total of four r(CGG)90-expressing mice compared with four WT controls was calculated. Error bars were used to indicate scanning electron microscopy (SEM), and the student's t-test was used to compare positively staining Purkinje neurons between r(CGG)90 and WT.
Drosophila genetics
Transgenic flies expressing rCGG90 repeats were described previously. UAS-TDP-43 transgenic lines were described previously (47). UAS-dsRNA-TBPH transgenic lines were obtained from Vienna Drosophila RNAi Center, Austria. All crosses were grown on standard medium at 25°C in 50–70% humidity.
Drosophila scanning electron microscopy
For SEM images, whole flies were dehydrated in gradient concentration ethanol (25, 50, 75 and 100%), dried with hexamethyldisilazane (Sigma), coated by electric field and argon gas and analyzed with an ISI DS-130 LaB6 SEM/STEM microscope.
Locomotion assay
Elav-Gal4; UAS-CGG60-EGFP transgenic flies (48 h of age) were collected for locomotor activity assays. To continuously monitor the locomotion, a DAM system (TriKinetics) was used. The readings were recorded each time the fly crossed the infrared beam. These interruptions were collected as a bin of 1 h. The activity of flies was calculated as the average number of movements in a given time recorded together with control flies. Data were collected and analyzed for locomotor activity.
Statistical methods
We used DAM Scan software (TriKinetics) to detect any outliers in the readings obtained. The readings obtained in a bin of 1 h were averaged over 24 h. Analyses to show significant differences between genotypes and treatments were performed using analysis of variance, post hoc t-tests (two-sample assuming equal variances). All data are shown as mean with standard error of mean.
Accession number
The microarray data reported in this paper have been deposited into the Gene Expression Omnibus database using the accession number GSE57034.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by the FRAXA Research Foundation and NIH grants HD24064, NS051630 and NS67461.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Nathaniel Heintz and Huda Zogbhi for providing us the Pcp2 BAC transgenic mice, Fen-Biao Gao for UAS-TDP-43 transgenic lines and Joseph Doyle, Joseph Dougherty and Myriam Heiman for discussion and useful suggestions. We also thank Dr Jeff Neul for the use of the motor-driven glass homogenizer as well as Dr Yanghong Gu and Debra Townley for technical advice. We also thank the following core facilities at Baylor College of Medicine: the Genomic and RNA Profiling Core, the NICHD Intellectual and Developmental Disabilities Research Center Neuropathology Core (HD24064) and the Integrated Microscopy Core.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 2.Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W., et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- 3.Brunberg J.A., Jacquemont S., Hagerman R.J., Berry-Kravis E.M., Grigsby J., Leehey M.A., Tassone F., Brown W.T., Greco C.M., Hagerman P.J. Fragile X premutation carriers: characteristic MR imaging findings of adult male patients with progressive cerebellar and cognitive dysfunction. Am. J. Neuroradiol. 2002;23:1757–1766. [PMC free article] [PubMed] [Google Scholar]
- 4.Greco C.M., Berman R.F., Martin R.M., Tassone F., Schwartz P.H., Chang A., Trapp B.D., Iwahashi C., Brunberg J., Grigsby J., et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 5.Greco C.M., Hagerman R.J., Tassone F., Chudley A.E., Del Bigio M.R., Jacquemont S., Leehey M., Hagerman P.J. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 6.Sellier C., Rau F., Liu Y., Tassone F., Hukema R.K., Gattoni R., Schneider A., Richard S., Willemsen R., Elliott D.J., et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tassone F., Iwahashi C., Hagerman P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 8.Udd B., Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- 9.Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Jin P. RNA-mediated neurodegeneration in fragile X-associated tremor/ataxia syndrome. Brain Res. 2012;1462:112–117. doi: 10.1016/j.brainres.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin P., Duan R., Qurashi A., Qin Y., Tian D., Rosser T.C., Liu H., Feng Y., Warren S.T. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 13.Sofola O.A., Jin P., Qin Y., Duan R., Liu H., de Haro M., Nelson D.L., Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qurashi A., Li W., Zhou J.Y., Peng J., Jin P. Nuclear accumulation of stress response mRNAs contributes to the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2011;7:e1002102. doi: 10.1371/journal.pgen.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sellier C., Freyermuth F., Tabet R., Tran T., He F., Ruffenach F., Alunni V., Moine H., Thibault C., Page A., et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashem V., Galloway J.N., Mori M., Willemsen R., Oostra B.A., Paylor R., Nelson D.L. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum. Mol. Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suarez-Farinas M., Schwarz C., Stephan D.A., Surmeier D.J., et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baloh R.H. How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other? Curr. Opin. Neurol. 2012;25:701–707. doi: 10.1097/WCO.0b013e32835a269b. [DOI] [PubMed] [Google Scholar]
- 19.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 20.Huang da W., Sherman B.T., Zheng X., Yang J., Imamichi T., Stephens R., Lempicki R.A. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinform. 2009 doi: 10.1002/0471250953.bi1311s27. 27:13.11.1-13.11.13. [DOI] [PubMed] [Google Scholar]
- 21.Qurashi A., Liu H., Ray L., Nelson D.L., Duan R., Jin P. Chemical screen reveals small molecules suppressing fragile X premutation rCGG repeat-mediated neurodegeneration in Drosophila. Hum. Mol. Genet. 2012;21:2068–2075. doi: 10.1093/hmg/dds024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornish K.M., Hocking D.R., Moss S.A., Kogan C.S. Selective executive markers of at-risk profiles associated with the fragile X premutation. Neurology. 2011;77:618–622. doi: 10.1212/WNL.0b013e3182299e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry-Kravis E., Hall D.A. Executive dysfunction in young FMR1 premutation carriers forme fruste of FXTAS or new phenotype? Neurology. 2011;77:612–613. doi: 10.1212/WNL.0b013e3182299f98. [DOI] [PubMed] [Google Scholar]
- 24.Moore C.J., Daly E.M., Schmitz N., Tassone F., Tysoe C., Hagerman R.J., Hagerman P.J., Morris R.G., Murphy K.C., Murphy D.G. A neuropsychological investigation of male premutation carriers of fragile X syndrome. Neuropsychologia. 2004;42:1934–1947. doi: 10.1016/j.neuropsychologia.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Kogan C.S., Turk J., Hagerman R.J., Cornish K.M. Impact of the Fragile X mental retardation 1 (FMR1) gene premutation on neuropsychiatric functioning in adult males without fragile X-associated Tremor/Ataxia syndrome: a controlled study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:859–872. doi: 10.1002/ajmg.b.30685. [DOI] [PubMed] [Google Scholar]
- 26.Roberts J.E., Bailey D.B., Jr., Mankowski J., Ford A., Sideris J., Weisenfeld L.A., Heath T.M., Golden R.N. Mood and anxiety disorders in females with the FMR1 premutation. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:130–139. doi: 10.1002/ajmg.b.30786. [DOI] [PubMed] [Google Scholar]
- 27.Aziz M., Stathopulu E., Callias M., Taylor C., Turk J., Oostra B., Willemsen R., Patton M. Clinical features of boys with fragile X premutations and intermediate alleles. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;121B:119–127. doi: 10.1002/ajmg.b.20030. [DOI] [PubMed] [Google Scholar]
- 28.Farzin F., Perry H., Hessl D., Loesch D., Cohen J., Bacalman S., Gane L., Tassone F., Hagerman P., Hagerman R. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J. Dev. Behav. Pediatr. 2006;27:S137–S144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 29.Goodlin-Jones B.L., Tassone F., Gane L.W., Hagerman R.J. Autistic spectrum disorder and the fragile X premutation. J. Dev. Behav. Pediatr. 2004;25:392–398. doi: 10.1097/00004703-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Cunningham C.L., Martinez Cerdeno V., Navarro Porras E., Prakash A.N., Angelastro J.M., Willemsen R., Hagerman P.J., Pessah I.N., Berman R.F., Noctor S.C. Premutation CGG-repeat expansion of the Fmr1 gene impairs mouse neocortical development. Hum. Mol. Genet. 2011;20:64–79. doi: 10.1093/hmg/ddq432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Tassone F., Berman R.F., Hagerman P.J., Hagerman R.J., Willemsen R., Pessah I.N. Murine hippocampal neurons expressing Fmr1 gene premutations show early developmental deficits and late degeneration. Hum. Mol. Genet. 2010;19:196–208. doi: 10.1093/hmg/ddp479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diep A.A., Hunsaker M.R., Kwock R., Kim K., Willemsen R., Berman R.F. Female CGG knock-in mice modeling the fragile X premutation are impaired on a skilled forelimb reaching task. Neurobiol. Learn. Mem. 2012;97:229–234. doi: 10.1016/j.nlm.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arocena D.G., Iwahashi C.K., Won N., Beilina A., Ludwig A.L., Tassone F., Schwartz P.H., Hagerman P.J. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum. Mol. Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- 34.Iwahashi C.K., Yasui D.H., An H.J., Greco C.M., Tassone F., Nannen K., Babineau B., Lebrilla C.B., Hagerman R.J., Hagerman P.J. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Arocena D., Yang J.E., Brouwer J.R., Tassone F., Iwahashi C., Berry-Kravis E.M., Goetz C.G., Sumis A.M., Zhou L., Nguyen D.V., et al. Fibroblast phenotype in male carriers of FMR1 premutation alleles. Hum. Mol. Genet. 2010;19:299–312. doi: 10.1093/hmg/ddp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Lee J.E., Wilusz J., Wilusz C.J. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J. Biol. Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moraes K.C., Wilusz C.J., Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 39.Brandmeir N.J., Geser F., Kwong L.K., Zimmerman E., Qian J., Lee V.M., Trojanowski J.Q. Severe subcortical TDP-43 pathology in sporadic frontotemporal lobar degeneration with motor neuron disease. Acta. Neuropathol. 2008;115:123–131. doi: 10.1007/s00401-007-0315-5. [DOI] [PubMed] [Google Scholar]
- 40.Giordana M.T., Piccinini M., Grifoni S., De Marco G., Vercellino M., Magistrello M., Pellerino A., Buccinna B., Lupino E., Rinaudo M.T. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol. 2010;20:351–360. doi: 10.1111/j.1750-3639.2009.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori F., Tanji K., Zhang H.X., Nishihira Y., Tan C.F., Takahashi H., Wakabayashi K. Maturation process of TDP-43-positive neuronal cytoplasmic inclusions in amyotrophic lateral sclerosis with and without dementia. Acta. Neuropathol. 2008;116:193–203. doi: 10.1007/s00401-008-0396-9. [DOI] [PubMed] [Google Scholar]
- 42.Strong M.J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Pamphlett R., Luquin N., McLean C., Jew S.K., Adams L. TDP-43 neuropathology is similar in sporadic amyotrophic lateral sclerosis with or without TDP-43 mutations. Neuropathol. Appl. Neurobiol. 2009;35:222–225. doi: 10.1111/j.1365-2990.2008.00982.x. [DOI] [PubMed] [Google Scholar]
- 44.Alami N.H., Smith R.B., Carrasco M.A., Williams L.A., Winborn C.S., Han S.S., Kiskinis E., Winborn B., Freibaum B.D., Kanagaraj A., et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C., et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin L.X., Beyer R.P., Hudson F.N., Linford N.J., Morris D.E., Kerr K.F. Evaluation of methods for oligonucleotide array data via quantitative real-time PCR. BMC Bioinformatics. 2006;7:23. doi: 10.1186/1471-2105-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y., Ferris J., Gao F.B. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol. Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.